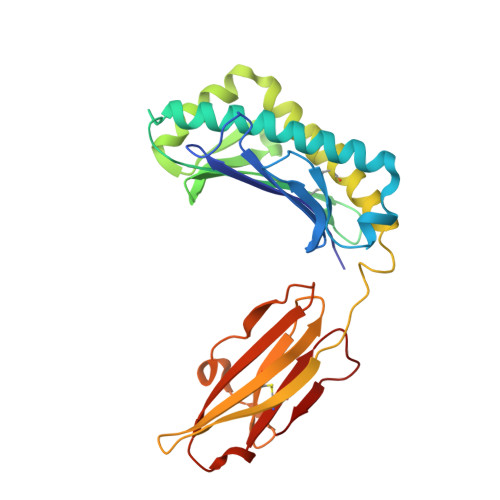
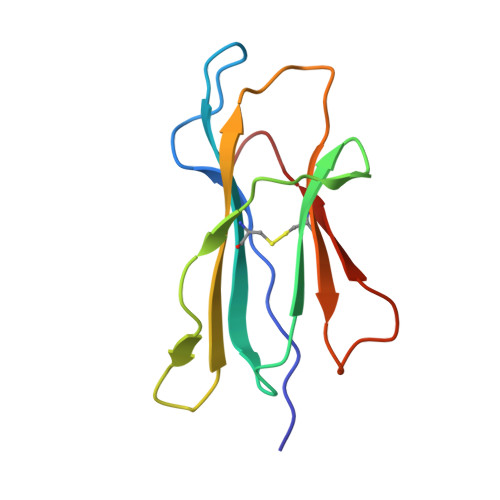
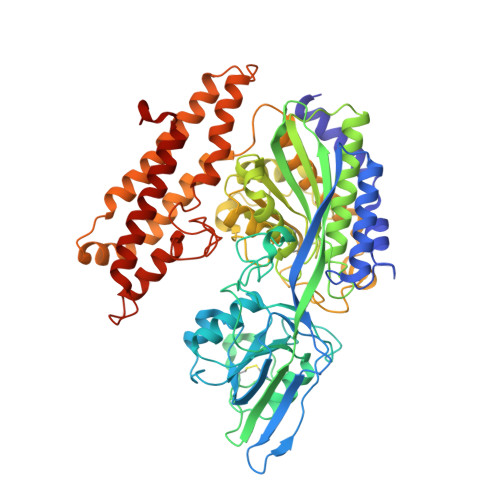
Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor.
Bennett, M.J., Lebron, J.A., Bjorkman, P.J.(2000) Nature 403: 46-53
- PubMed: 10638746
- DOI: https://doi.org/10.1038/47417
- Primary Citation of Related Structures:
1DE4 - PubMed Abstract:
HFE is related to major histocompatibility complex (MHC) class I proteins and is mutated in the iron-overload disease hereditary haemochromatosis. HFE binds to the transferrin receptor (TfR), a receptor by which cells acquire iron-loaded transferrin. The 2.8 A crystal structure of a complex between the extracellular portions of HFE and TfR shows two HFE molecules which grasp each side of a twofold symmetric TfR dimer. On a cell membrane containing both proteins, HFE would 'lie down' parallel to the membrane, such that the HFE helices that delineate the counterpart of the MHC peptide-binding groove make extensive contacts with helices in the TfR dimerization domain. The structures of TfR alone and complexed with HFE differ in their domain arrangement and dimer interfaces, providing a mechanism for communicating binding events between TfR chains. The HFE-TfR complex suggests a binding site for transferrin on TfR and sheds light upon the function of HFE in regulating iron homeostasis.
- Division of Biology, California Institute of Technology, Pasadena 91125, USA.
Organizational Affiliation: