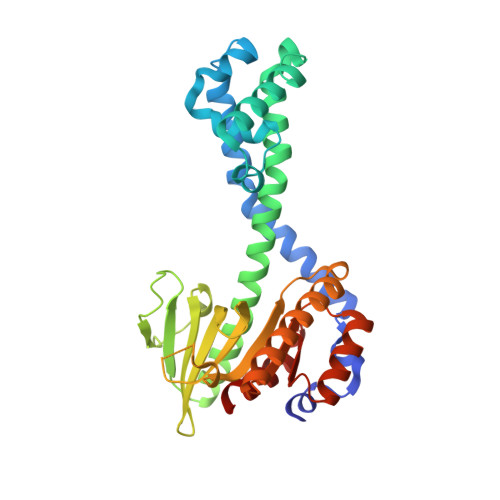
Restriction enzyme BsoBI-DNA complex: a tunnel for recognition of degenerate DNA sequences and potential histidine catalysis.
van der Woerd, M.J., Pelletier, J.J., Xu, S., Friedman, A.M.(2001) Structure 9: 133-144
- PubMed: 11250198
- DOI: https://doi.org/10.1016/s0969-2126(01)00564-0
- Primary Citation of Related Structures:
1DC1 - PubMed Abstract:
Restriction endonucleases form a diverse family of proteins with substantial variation in sequence, structure, and interaction with recognition site DNA. BsoBI is a thermophilic restriction endonuclease that exhibits both base-specific and degenerate recognition within the sequence CPyCGPuG. The structure of BsoBI complexed to cognate DNA has been determined to 1.7 A resolution, revealing several unprecedented features. Each BsoBI monomer is formed by inserting a helical domain into an expanded EcoRI-type catalytic domain. DNA is completely encircled by a BsoBI dimer. Recognition sequence DNA lies within a 20 A long tunnel of protein that excludes bulk solvent. Interactions with the specific bases are made in both grooves through direct and water-mediated hydrogen bonding. Interaction with the degenerate position is mediated by a purine-specific hydrogen bond to N7, ensuring specificity, and water-mediated H bonding to the purine N6/O6 and pyrimidine N4/O4, allowing degeneracy. In addition to the conserved active site residues of the DX(n)(E/D)ZK restriction enzyme motif, His253 is positioned to act as a general base. A catalytic mechanism employing His253 and two metal ions is proposed. If confirmed, this would be the first example of histidine-mediated catalysis in a restriction endonuclease. The structure also provides two novel examples of the role of water in protein-DNA interaction. Degenerate recognition may be mediated by employing water as a hydrogen bond donor or acceptor. The structure of DNA in the tunnel may also be influenced by the absence of bulk solvent.
- Department of Biological Sciences, Purdue University, 47907, West Lafayette, IN, USA.
Organizational Affiliation: