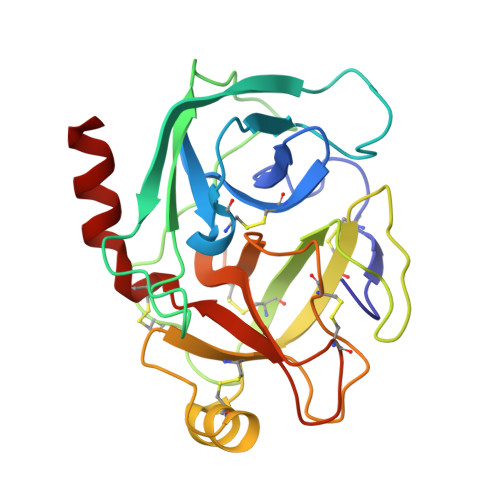
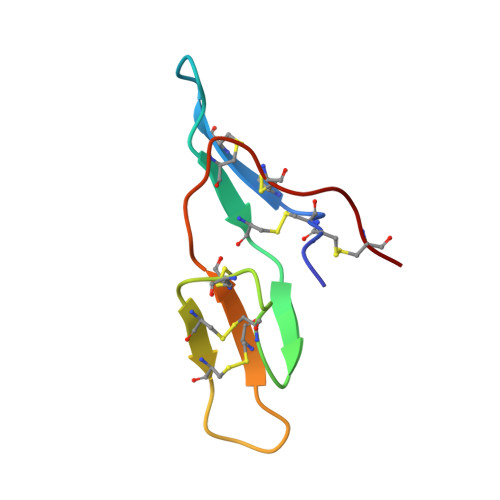
Crystal structure of cancer chemopreventive Bowman-Birk inhibitor in ternary complex with bovine trypsin at 2.3 A resolution. Structural basis of Janus-faced serine protease inhibitor specificity.
Koepke, J., Ermler, U., Warkentin, E., Wenzl, G., Flecker, P.(2000) J Mol Biology 298: 477-491
- PubMed: 10772864
- DOI: https://doi.org/10.1006/jmbi.2000.3677
- Primary Citation of Related Structures:
1D6R - PubMed Abstract:
Understanding molecular recognition on a structural basis is an objective with broad academic and applied significance. In the complexes of serine proteases and their proteinaceous inhibitors, recognition is governed mainly by residue P1 in accord with primary serine protease specificity. The bifunctional soybean Bowman-Birk inhibitor (sBBI) should, therefore, interact at LysI16 (subdomain 1) with trypsin and at LeuI43 (subdomain 2) with chymotrypsin. In contrast with this prediction, a 2:1 assembly with trypsin was observed in solution and in the crystal structure of sBBI in complex with trypsin, determined at 2.3 A resolution by molecular replacement. Strikingly, P1LeuI43 of sBBI was fully embedded into the S(1) pocket of trypsin in contrast to primary specificity. The triple-stranded beta-hairpin unique to the BBI-family and the surface loops surrounding the active site of the enzyme formed a protein-protein-interface far extended beyond the primary contact region. Polar residues, hydrophilic bridges and weak hydrophobic contacts were predominant in subdomain 1, interacting specifically with trypsin. However, close hydrophobic contacts across the interface were characteristic of subdomain 2 reacting with both trypsin and chymotrypsin. A Met27Ile replacement shifted the ratio with trypsin to the predicted 1:1 ratio. Thus, the buried salt-bridge responsible for trypsin specificity was stabilised in a polar, and destabilized in a hydrophobic, environment. This may be used for adjusting the specificity of protease inhibitors for applications such as insecticides and cancer chemopreventive agents.
- Max Planck Institut für Biophysik, Frankfurt am Main, Germany.
Organizational Affiliation: