The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
Tanaka, N., Yamane, T., Tsukihara, T., Ashida, T., Kakudo, M.(1975) J Biochem 77: 147-162
- PubMed: 166072
- Primary Citation of Related Structures:
1CYC - PubMed Abstract:
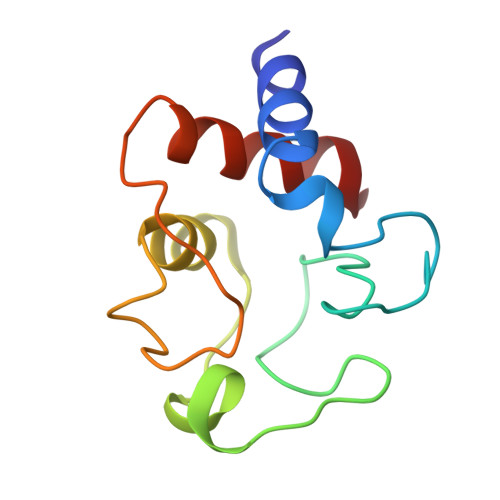
The structure analysis of bonito heart ferrocytochrome c was carried out at 2.3 A resolution by X-ray diffraction, and a Kendrew-type skeletal model was built up. This molecule has an overall egg shape, 35 A in height, 30 A in width and 23 A in thickness; the 5th ligand of the heme iron atom is the N-epsilon atom of the His-18 imidazole ring and the 6th is the Met-80 sulfur atom. Distinct alpha-helix regions are found between the N-terminus and reside 11, between 60 and 69, and between 90 and the C-terminus. The most distinct difference between the conformation of the present molecule and that of the horse oxidized molecule is the location of the Phe-82 phenyl ring. In the present reduced molecule, the phyenyl ring is in closer contact with the iron atom and gives influences on the character of the iron atom. Inside the molecule, at the lower part of the heme pocket, there is an extended hydrogen bond network including the propionic acid residues of the heme group. Both Phe-82 and the hydrogen bond network may play a key role in the function of this molecule.