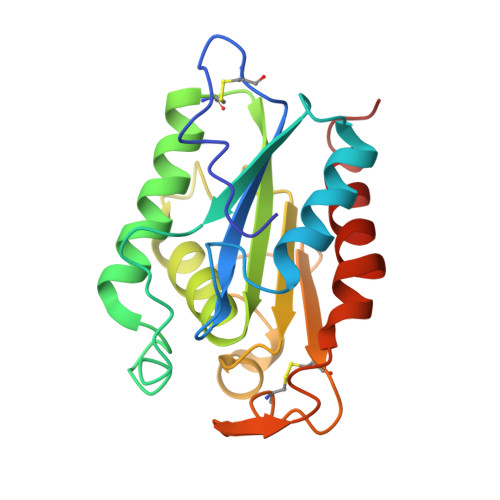
Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent.
Martinez, C., De Geus, P., Lauwereys, M., Matthyssens, G., Cambillau, C.(1992) Nature 356: 615-618
- PubMed: 1560844
- DOI: https://doi.org/10.1038/356615a0
- Primary Citation of Related Structures:
1CUS - PubMed Abstract:
Lipases belong to a class of esterases whose activity on triglycerides is greatly enhanced at lipid-water interfaces. This phenomenon, called interfacial activation, has a structural explanation: a hydrophobic lid, which at rest covers the catalytic site, is displaced on substrate or inhibitor binding and probably interacts with the lipid matrix. Fusarium solani pisi cutinase belongs to a group of homologous enzymes of relative molecular mass 22-25K (ref. 7) capable of degrading cutin, the insoluble lipid-polyester matrix covering the surface of plants, and hydrolysing triglycerides. Cutinases differ from classical lipases in that they do not exhibit interfacial activation; they are active on soluble as well as on emulsified triglycerides. Cutinases therefore establish a bridge between esterases and lipases. We report here the three-dimensional structure of a recombinant cutinase from F. solani pisi, expressed in Escherichia coli. Cutinase is an alpha-beta protein; the active site is composed of the triad Ser 120, His 188 and Asp 175. Unlike other lipases, the catalytic serine is not buried under surface loops, but is accessible to solvent. This could explain why cutinase does not display interfacial activation.
- Faculté de médecine Nord, France.
Organizational Affiliation: