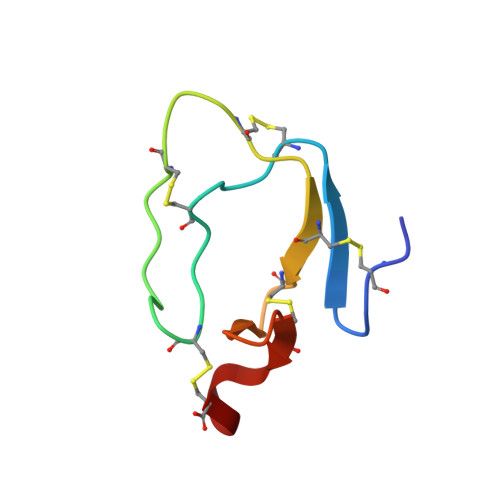
NMR solution structure of Apis mellifera chymotrypsin/cathepsin G inhibitor-1 (AMCI-1): structural similarity with Ascaris protease inhibitors
Cierpicki, T., Bania, J., Otlewski, J.(2000) Protein Sci 9: 976-984
- PubMed: 10850807
- DOI: https://doi.org/10.1110/ps.9.5.976
- Primary Citation of Related Structures:
1CCV - PubMed Abstract:
The three-dimensional structure of the 56 residue polypeptide Apis mellifera chymotrypsin/cathepsin G inhibitor 1 (AMCI-1) isolated from honey bee hemolymph was calculated based on 730 experimental NMR restraints. It consists of two approximately perpendicular beta-sheets, several turns, and a long exposed loop that includes the protease binding site. The lack of extensive secondary structure features or hydrophobic core is compensated by the presence of five disulfide bridges that stabilize both the protein scaffold and the binding loop segment. A detailed analysis of the protease binding loop conformation reveals that it is similar to those found in other canonical serine protease inhibitors. The AMCI-1 structure exhibits a common fold with a novel family of inhibitors from the intestinal parasitic worm Ascaris suum. The pH-induced conformational changes in the binding loop region observed in the Ascaris inhibitor ATI are absent in AMCI-1. Similar binding site sequences and structures strongly suggest that the lack of the conformational change can be attributed to a Glu-->Gln substitution at the P1' position in AMCI-1, compared to ATI. Analysis of amide proton temperature coefficients shows very good correlation with the presence of hydrogen bond donors in the calculated AMCI-1 structure.
- Institute of Biochemistry and Molecular Biology, University of Wroclaw, Poland.
Organizational Affiliation: