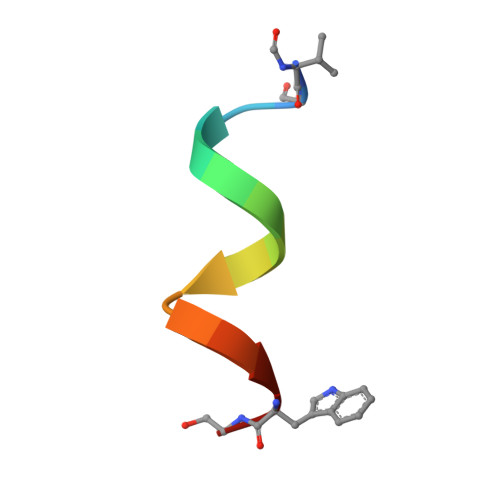
The Gramicidin Pore: Crystal Structure of a Cesium Complex.
Wallace, B.A., Ravikumar, K.(1988) Science 241: 182
- PubMed: 2455344
- DOI: https://doi.org/10.1126/science.2455344
- Primary Citation of Related Structures:
1C4D - PubMed Abstract:
Gramicidin, a linear polypeptide composed of hydrophobic amino acids with alternating L- and D- configurations, forms transmembrane ion channels. The crystal structure of a gramicidin-cesium complex has been determined at 2.0 angstrom resolution. In this structure, gramicidin forms a 26 angstrom long tube comprised of two polypeptide chains arranged as antiparallel beta strands that are wrapped into a left-handed helical coil with 6.4 residues per turn. The polypeptide backbone forms the interior of the hydrophilic, solvent-filled pore and the side chains form a hydrophobic and relatively regular surface on the outside of the pore. This example of a crystal structure of a solvent-filled ion pore provides a basis for understanding the physical nature of ion translocation.
- Department of Chemistry and Center for Biophysics, Rensselaer Polytechnic Institute, Troy, NY 12180.
Organizational Affiliation: