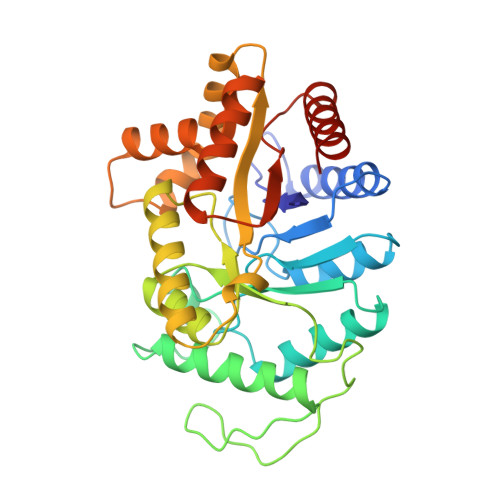
Structure of the beta 2 homodimer of bacterial luciferase from Vibrio harveyi: X-ray analysis of a kinetic protein folding trap.
Thoden, J.B., Holden, H.M., Fisher, A.J., Sinclair, J.F., Wesenberg, G., Baldwin, T.O., Rayment, I.(1997) Protein Sci 6: 13-23
- PubMed: 9007973
- DOI: https://doi.org/10.1002/pro.5560060103
- Primary Citation of Related Structures:
1BSL - PubMed Abstract:
Luciferase, as isolated from Vibrio harveyi, is an alpha beta heterodimer. When allowed to fold in the absence of the alpha subunit, either in vitro or in vivo, the beta subunit of enzyme will form a kinetically stable homodimer that does not unfold even after prolonged incubation in 5 M urea at pH 7.0 and 18 degrees C. This form of the beta subunit, arising via kinetic partitioning on the folding pathway, appears to constitute a kinetically trapped alternative to the heterodimeric enzyme (Sinclair JF, Ziegler MM, Baldwin TO. 1994. Kinetic partitioning during protein folding yields multiple native states. Nature Struct Biol 1: 320-326). Here we describe the X-ray crystal structure of the beta 2 homodimer of luciferase from V. harveyi determined and refined at 1.95 A resolution. Crystals employed in the investigational belonged to the orthorhombic space group P2(1)2(1)2(1) with unit cell dimensions of a = 58.8 A, b = 62.0 A, and c = 218.2 A and contained one dimer per asymmetric unit. Like that observed in the functional luciferase alpha beta heterodimer, the major tertiary structural motif of each beta subunit consists of an (alpha/beta)8 barrel (Fisher AJ, Raushel FM, Baldwin TO, Rayment I. 1995. Three-dimensional structure of bacterial luciferase from Vibrio harveyi at 2.4 A resolution. Biochemistry 34: 6581-6586). The root-mean-square deviation of the alpha-carbon coordinates between the beta subunits of the hetero- and homodimers is 0.7 A. This high resolution X-ray analysis demonstrated that "domain" or "loop" swapping has not occurred upon formation of the beta 2 homodimer and thus the stability of the beta 2 species to denaturation cannot be explained in such simple terms. In fact, the subunit:subunit interfaces observed in both the beta 2 homodimer and alpha beta heterodimer are remarkably similar in hydrogen-bonding patterns and buried surface areas.
- Institute for Enzyme Research, University of Wisconsin, Madison 53705, USA.
Organizational Affiliation: