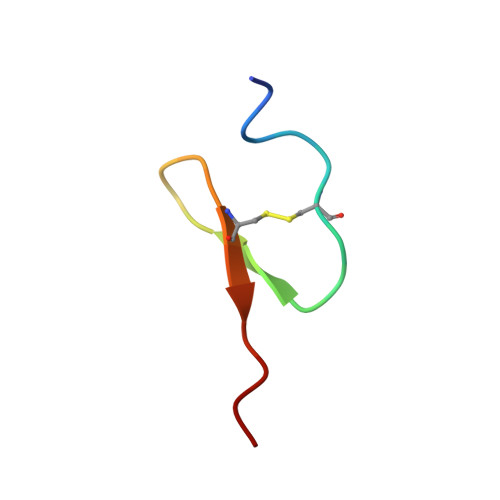
Solution structure of an insect growth factor, growth-blocking peptide.
Aizawa, T., Fujitani, N., Hayakawa, Y., Ohnishi, A., Ohkubo, T., Kumaki, Y., Kawano, K., Hikichi, K., Nitta, K.(1999) J Biological Chem 274: 1887-1890
- PubMed: 9890941
- DOI: https://doi.org/10.1074/jbc.274.4.1887
- Primary Citation of Related Structures:
1BQF - PubMed Abstract:
Growth-blocking peptide (GBP) is an insect growth factor consisting of 25 amino acid residues that retards the development of lepidopteran larvae at high concentration while it stimulates larval growth at low concentration. In this study, we determined the solution structure of GBP by two-dimensional 1H NMR spectroscopy. The structure contains a short segment of double-stranded beta-sheet involving residues 11-13 and 19-21 and a type-II beta-turn in the loop region (residues 8-11), whereas the N and C termini are disordered. This is the first report of the three-dimensional structure of the peptiderigic insect growth factor, and the structure of the well defined region of GBP was found to share similarity with that of the C-terminal domain of the epidermal growth factor (EGF). Because GBP has been reported to stimulate DNA synthesis of not only insect cells but also human keratinocyte cells at the same level with EGF, the structural similarity between GBP and EGF may lead to the interaction of GBP to EGF receptor.
- Division of Biological Sciences, Graduate School of Science, Hokkaido University, Sapporo 060-0810, Japan.
Organizational Affiliation: