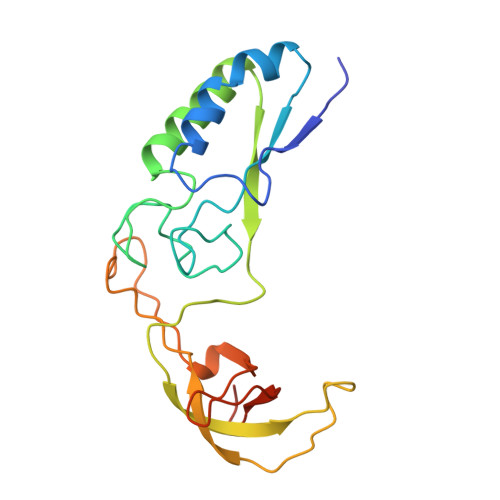
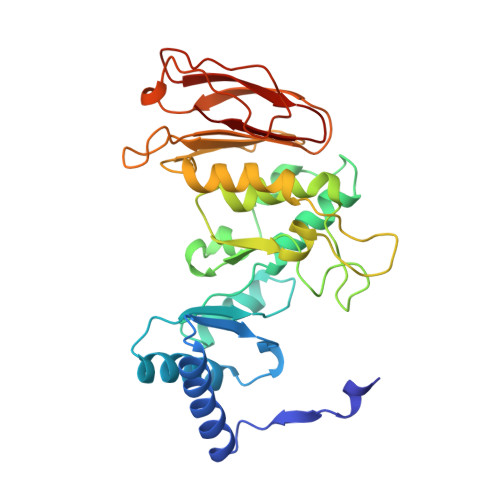
Crystal structure of calcium-depleted Bacillus licheniformis alpha-amylase at 2.2 A resolution.
Machius, M., Wiegand, G., Huber, R.(1995) J Mol Biology 246: 545-559
- PubMed: 7877175
- DOI: https://doi.org/10.1006/jmbi.1994.0106
- Primary Citation of Related Structures:
1BPL - PubMed Abstract:
The three-dimensional structure of the calcium-free form of Bacillus licheniformis alpha-amylase (BLA) has been determined by multiple isomorphous replacement in a crystal of space group P4(3)2(1)2 (a = b = 119.6 A, c = 85.4 A). The structure was refined using restrained crystallographic refinement to an R-factor of 0.177 for 28,147 independent reflections with intensities FObs > 0 at 2.2 A resolution, with root mean square deviations of 0.008 A and 1.4 degrees from ideal bond lengths and bond angles, respectively. The final model contains 469 residue, 237 water molecules, and one chloride ion. The segment between Trp182 and Asn192 could not be located in the electron density, nor could the N and C termini. Cleavage of the calcium-free form of BLA was observed after Glu189, due to a Glu-C endopeptidase present in trace amounts in the preparation. BLA did not crystallize without this cleavage under the conditions applied. BLA exhibits the characteristic overall topological fold observed for other alpha-amylases and related amylolytic enzymes: a central domain A containing an alpha/beta-barrel with a large protrusion between beta-strand 3 and alpha-helix 3 (domain B) and a C-terminal greek key motif (domain C). Unlike in the other enzymes, domain B possesses a beta-sheet made up of six loosely connected, twisted beta-strands forming a kind of a barrel with a large hole in the interior. Topological comparisons to TAKA-amylase, pig pancreatic alpha-amylase and cyclodextrin glycosyltransferase reveal a very high structural equivalence for large portions of the proteins and an exceptionally pronounced structural similarity for calcium binding, chloride binding and the active site. None of the theories proposed to explain the enhanced thermostability of BLA showed a satisfactory correlation with the three-dimensional structure. Instead, sequence comparisons to the less thermostable bacterial alpha-amylase from Bacillus amyloliquefaciens (BAA) indicate that some ionic interactions present in BLA, but which cannot be formed in BAA, might be responsible for the enhanced thermostability of BLA.
- Max-Planck-Institut für Biochemie, Planegg-Martinsried, Germany.
Organizational Affiliation: