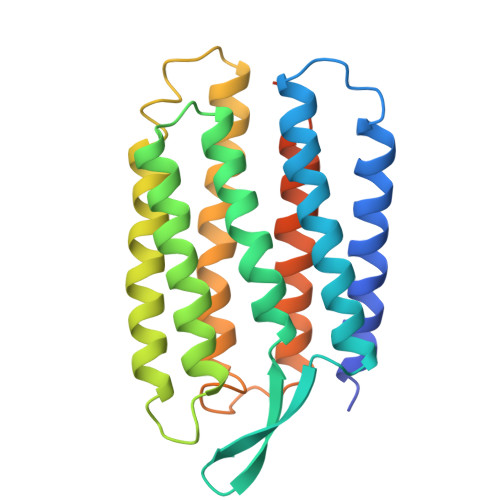
Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Sato, H., Takeda, K., Tani, K., Hino, T., Okada, T., Nakasako, M., Kamiya, N., Kouyama, T.(1999) Acta Crystallogr D Biol Crystallogr 55: 1251-1256
- PubMed: 10393291
- DOI: https://doi.org/10.1107/s090744499900503x
- Primary Citation of Related Structures:
1BM1 - PubMed Abstract:
In the purple membrane of Halobacterium salinarium, bacteriorhodopsin trimers are arranged in a hexagonal lattice. When purple membrane sheets are incubated at high temperature with neutral detergent, membrane vesicularization takes place, yielding inside-out vesicles with a diameter of 50 nm. The vesicular structure becomes unstable at low temperature, where successive fusion of the vesicles yields a crystal which is composed of stacked planar membranes. X-ray crystallographic analysis reveals that the bacteriorhodopsin trimers are arranged in a honeycomb lattice in each membrane layer and that neighbouring membranes orient in opposite directions. The native structure of the trimeric unit is preserved in the honeycomb lattice, irrespective of alterations in the in-plane orientation of the trimer. One phospholipid tightly bound to a crevice between monomers in the trimeric unit is suggested to act as a glue in the formation of the trimer.
- Department of Physics, Graduate School of Science, Nagoya University, Nagoya 464-8602, Japan.
Organizational Affiliation: