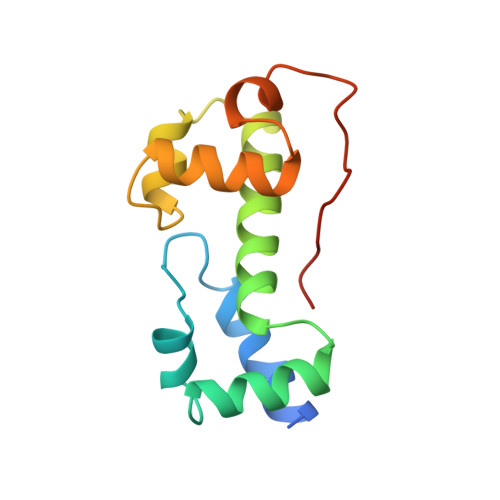
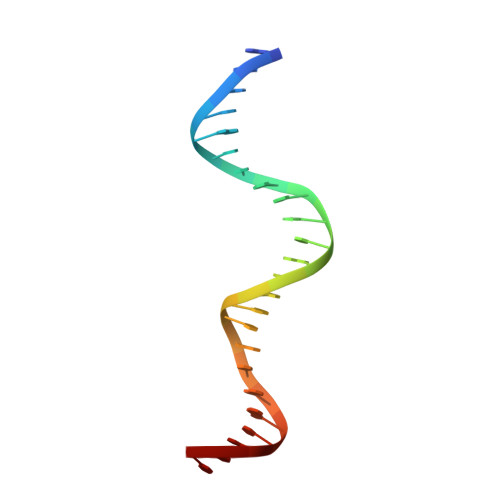
A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator.
Rhee, S., Martin, R.G., Rosner, J.L., Davies, D.R.(1998) Proc Natl Acad Sci U S A 95: 10413-10418
- PubMed: 9724717
- DOI: https://doi.org/10.1073/pnas.95.18.10413
- Primary Citation of Related Structures:
1BL0 - PubMed Abstract:
A crystal structure for a member of the AraC prokaryotic transcriptional activator family, MarA, in complex with its cognate DNA-binding site is described. MarA consists of two similar subdomains, each containing a helix-turn-helix DNA-binding motif. The two recognition helices of the motifs are inserted into adjacent major groove segments on the same face of the DNA but are separated by only 27 A thereby bending the DNA by approximately 35 degrees. Extensive interactions between the recognition helices and the DNA major groove provide the sequence specificity.
- Laboratory of Molecular Biology, National Institutes of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892-0560, USA.
Organizational Affiliation: