Solution structure and base pair opening kinetics of the i-motif dimer of d(5mCCTTTACC): a noncanonical structure with possible roles in chromosome stability.
Nonin, S., Phan, A.T., Leroy, J.L.(1997) Structure 5: 1231-1246
- PubMed: 9331414
- DOI: https://doi.org/10.1016/s0969-2126(97)00273-6
- Primary Citation of Related Structures:
1BAE - PubMed Abstract:
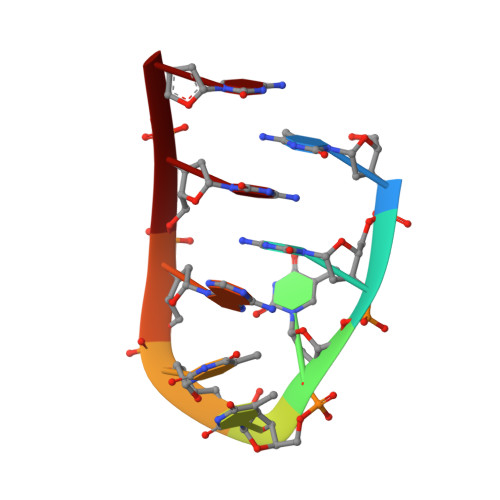
Repetitive cytosine-rich DNA sequences have been identified in telomeres and centromeres of eukaryotic chromosomes. These sequences play a role in maintaining chromosome stability during replication and may be involved in chromosome pairing during meiosis. The C-rich repeats can fold into an 'i-motif' structure, in which two parallel-stranded duplexes with hemiprotonated C.C+ pairs are intercalated. Previous NMR studies of naturally occurring repeats have produced poor NMR spectra. This led us to investigate oligonucleotides, based on natural sequences, to produce higher quality spectra and thus provide further information as to the structure and possible biological function of the i-motif. NMR spectroscopy has shown that d(5mCCTTTACC) forms an i-motif dimer of symmetry-related and intercalated folded strands. The high-definition structure is computed on the basis of the build-up rates of 29 intraresidue and 35 interresidue nuclear Overhauser effect (NOE) connectivities. The i-motif core includes intercalated interstrand C.C+ pairs stacked in the order 2*.8/1.7*/1*.7/2.8* (where one strand is distinguished by an asterisk and the numbers relate to the base positions within the repeat). The TTTA sequences form two loops which span the two wide grooves on opposite sides of the i-motif core; the i-motif core is extended at both ends by the stacking of A6 onto C2.C8+. The lifetimes of pairs C2.C8+ and 5mC1.C7+ are 1 ms and 1 s, respectively, at 15 degrees C. Anomalous exchange properties of the T3 imino proton indicate hydrogen bonding to A6 N7 via a water bridge. The d(5mCCTTTTCC) deoxyoligonucleotide, in which position 6 is occupied by a thymidine instead of an adenine, also forms a symmetric i-motif dimer. However, in this structure the two TTTT loops are located on the same side of the i-motif core and the C.C+ pairs are formed by equivalent cytidines stacked in the order 8*.8/1.1*/7*.7/2.2*. Oligodeoxynucleotides containing two C-rich repeats can fold and dimerize into an i-motif. The change of folding topology resulting from the substitution of a single nucleoside emphasizes the influence of the loop residues on the i-motif structure formed by two folded strands.
- Groupe de Biophysique, de l'Ecole Polytechnique et de l'URA, CNRS, Palaiseau, France.
Organizational Affiliation: