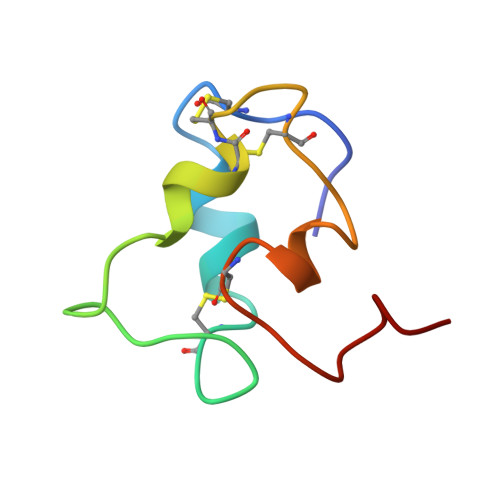
Solution structure of a mini IGF-1.
De Wolf, E., Gill, R., Geddes, S., Pitts, J., Wollmer, A., Grotzinger, J.(1996) Protein Sci 5: 2193-2202
- PubMed: 8931138
- DOI: https://doi.org/10.1002/pro.5560051106
- Primary Citation of Related Structures:
1B9G - PubMed Abstract:
Mini insulin-like growth factor 1, an inactive insulin-like growth factor 1 mutant lacking the C region, was studied by 2D NMR spectroscopy. Resonances were assigned for almost all protons of the 57 amino acid residues. The 3D structure of the protein was determined by distance geometry methods. Three helical segments; Ala 8-Cys 18, Gly 42-Phe 49, and Leu 54-Cys 61, were identified, corresponding to those present in wild-type insulin-like growth factor 1 and in single-chain insulin. Their relative orientation, however, was found to be changed. This change is connected with a displacement of the Phe 23-Tyr 24-Phe 25-Asn 26 beta-strand-like segment, i.e., of aromatic side chains known to be important for receptor binding. Thus, deletion of the C region of IGF-1 results in a substantial tertiary structural rearrangement that accounts for the loss of receptor affinity.
- Institut für Biochemie, Rheinisch-Westfälische Technische Hochsclule Aachen, Germany.
Organizational Affiliation: