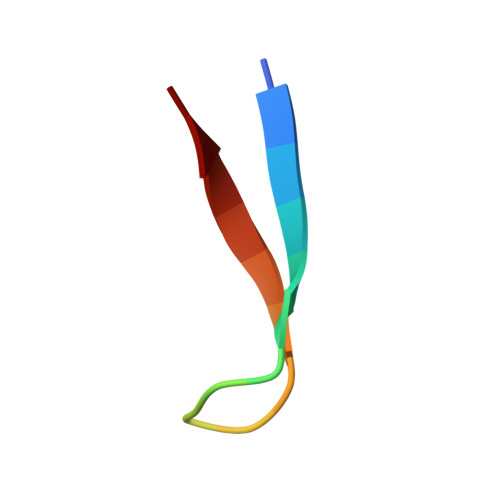
A cis proline turn linking two beta-hairpin strands in the solution structure of an antibody-bound HIV-1IIIB V3 peptide.
Tugarinov, V., Zvi, A., Levy, R., Anglister, J.(1999) Nat Struct Biol 6: 331-335
- PubMed: 10201400
- DOI: https://doi.org/10.1038/7567
- Primary Citation of Related Structures:
1B03 - PubMed Abstract:
The refined solution structure of an 18-residue HIV-1IIIB V3 peptide in complex with the Fv fragment of an anti-gp120 antibody reveals an unexpected type VI beta-turn comprising residues RGPG at the center of a beta-hairpin. The central glycine and proline of this turn are linked by a cis peptide bond. The residues of the turn interact extensively with the antibody Fv. 15N[1H] NOE measurements show that the backbone of the peptide, including the central QRGPGR loop, is well ordered in the complex. The solution structure is significantly different from the X-ray structures of HIV-1MN V3 peptides bound to anti-peptide antibodies. These differences could be due to a two-residue (QR) insertion preceding the GPGR sequence in the HIV-1IIIB strain, and the much longer peptide epitope immobilized by the anti-gp120 antibody.
- Department of Structural Biology, The Weizmann Institute of Science, Rehovot, Israel.
Organizational Affiliation: