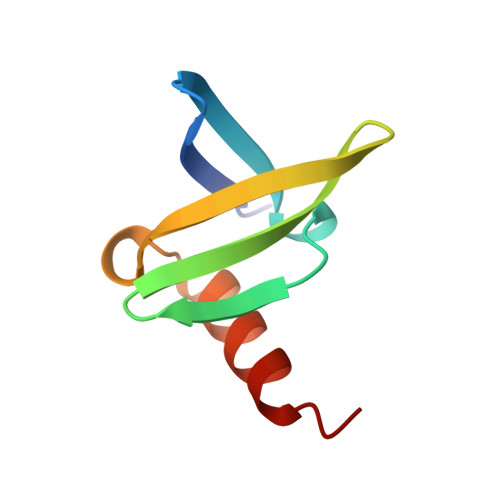
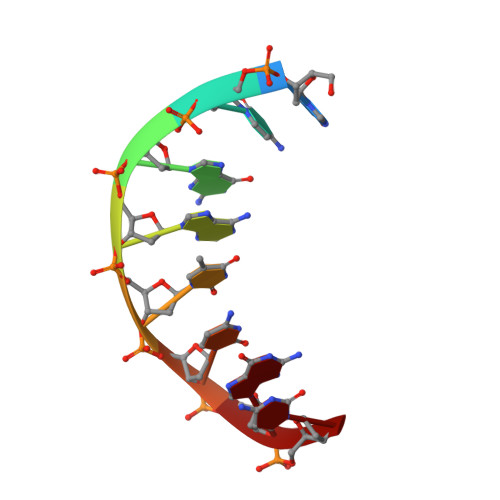
The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Robinson, H., Gao, Y.G., McCrary, B.S., Edmondson, S.P., Shriver, J.W., Wang, A.H.(1998) Nature 392: 202-205
- PubMed: 9515968
- DOI: https://doi.org/10.1038/32455
- Primary Citation of Related Structures:
1AZP, 1AZQ - PubMed Abstract:
The proteins Sac7d and Sso7d belong to a class of small chromosomal proteins from the hyperthermophilic archaeon Sulfolobus acidocaldarius and S. solfactaricus, respectively. These proteins are extremely stable to heat, acid and chemical agents. Sac7d binds to DNA without any particular sequence preference and thereby increases its melting temperature by approximately 40 degrees C. We have now solved and refined the crystal structure of Sac7d in complex with two DNA sequences to high resolution. The structures are examples of a nonspecific DNA-binding protein bound to DNA, and reveal that Sac7d binds in the minor groove, causing a sharp kinking of the DNA helix that is more marked than that induced by any sequence-specific DNA-binding proteins. The kink results from the intercalation of specific hydrophobic side chains of Sac7d into the DNA structure, but without causing any significant distortion of the protein structure relative to the uncomplexed protein in solution.
- Department of Cell and Structural Biology, University of Illinois at Urbana-Champaign, Urbana 61801, USA.
Organizational Affiliation: