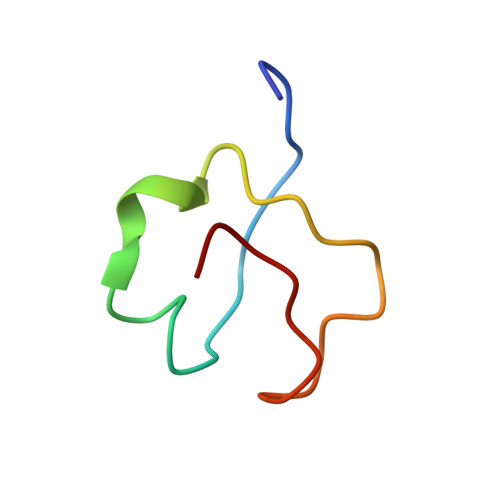
3D solution structure of copper and silver-substituted yeast metallothioneins.
Peterson, C.W., Narula, S.S., Armitage, I.M.(1996) FEBS Lett 379: 85-93
- PubMed: 8566237
- DOI: https://doi.org/10.1016/0014-5793(95)01492-6
- Primary Citation of Related Structures:
1AOO, 1AQQ, 1AQR, 1AQS - PubMed Abstract:
3D solution structural calculations for yeast silver(I)-substituted metallothionein (MT) and native copper(I) MT were completed using experimentally determined NOE and dihedral angle constraints, in conjunction with experimentally derived metal-to-Cys connectivities for AgMT which were assumed identical for CuMT. For the first 40 residues in both structures, the polypeptide backbone wraps around the metal cluster in two large parallel loops separated by a deep cleft containing the metal cluster. Minor differences between the two structures include differences in hydrogen bonds and the orientation of the N-terminus with the overall protein volume conserved to within 6.5%.
- Physics Department, Univ. of Connecticut, Storrs 06269-3046, USA.
Organizational Affiliation: