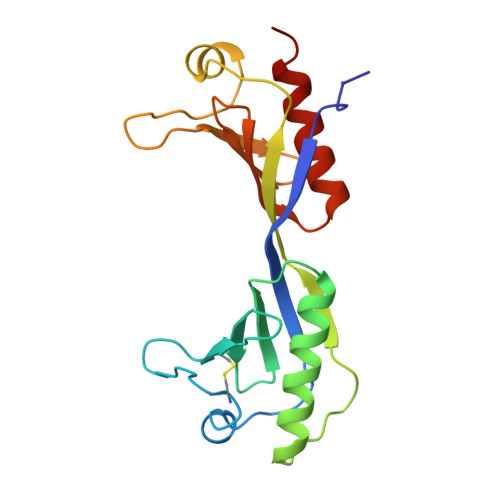
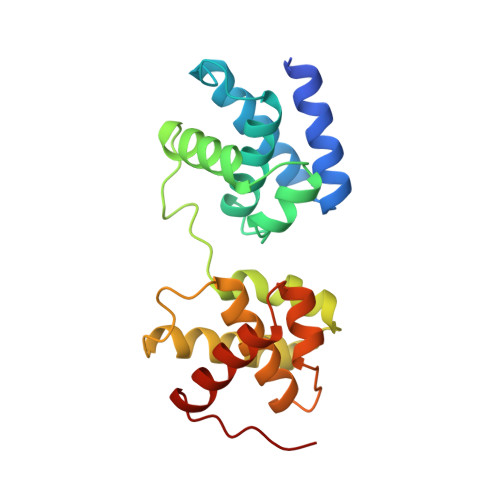
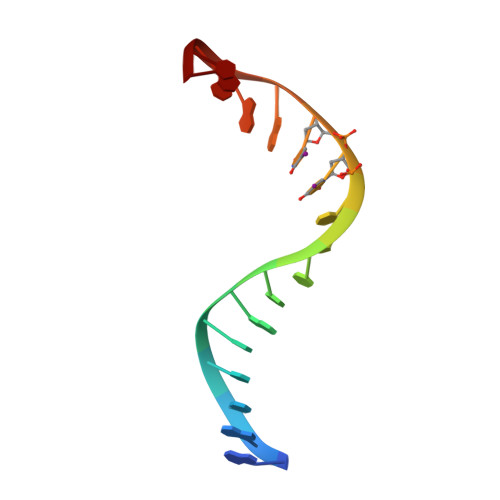
The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box.
Kosa, P.F., Ghosh, G., DeDecker, B.S., Sigler, P.B.(1997) Proc Natl Acad Sci U S A 94: 6042-6047
- PubMed: 9177165
- DOI: https://doi.org/10.1073/pnas.94.12.6042
- Primary Citation of Related Structures:
1AIS - PubMed Abstract:
Archaea possess a basal transcriptional apparatus that resembles that of eukaryotes. Here we report the 2.1-A crystal structure of the archaeal transcription factor complex formed by the TATA-box-binding protein (TBP), the transcription factor IIB homolog, and a DNA target, all from the hyperthermophile Pyrococcus woesei. The overall fold of these two basal transcription factors is essentially the same as that of their eukaryotic counterparts. However, in comparison with the eukaryotic complexes, the archaeal TBP-DNA interface is more symmetrical, and in this structure the orientation of the preinitiation complex assembly on the promoter is inverted with respect to that seen in all crystal structures of comparable eukaryotic systems. This study of the structural details of an archaeal transcription factor complex presents the opportunity to examine the evolution of the basal eukaryotic transcriptional apparatus from a stereochemical viewpoint and to extend our understanding of the physical biochemistry of transcriptional initiation.
- Department of Molecular Biophysics and Biochemistry, and the Howard Hughes Medical Institute, Yale University, 260 Whitney Avenue, JWG 423, New Haven CT 06511, USA.
Organizational Affiliation: