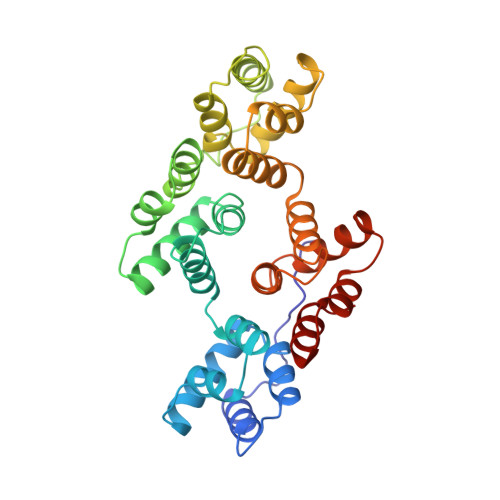
Crystal structure of the annexin XII hexamer and implications for bilayer insertion.
Luecke, H., Chang, B.T., Mailliard, W.S., Schlaepfer, D.D., Haigler, H.T.(1995) Nature 378: 512-515
- PubMed: 7477411
- DOI: https://doi.org/10.1038/378512a0
- Primary Citation of Related Structures:
1AEI - PubMed Abstract:
Annexins are a family of calcium- and phospholipid-binding proteins implicated in a number of biological processes including membrane fusion and ion channel formation. The crystal structure of the annexin XII hexamer, refined at 2.8 A resolution, forms a concave disk with 3-2 symmetry, about 100 A in diameter and 70 A thick with a central hydrophilic pore. Six intermolecular Ca2+ ions are involved in hexamer formation. An additional 18 Ca2+ ions are located on the perimeter of the disk, accessible only from the side of the hexameric disk. On the basis of the hexamer structure we propose here a new mode of protein-phospholipid bilayer interaction that is distinct from the hydrophobic insertion of typical membrane proteins. This speculative model postulates the Ca(2+)-dependent insertion of the hydrophilic annexin XII hexamer into phospholipid bilayers with local reorientation of the bilayer phospholipids.
- Stanford Synchrotron Radiation Laboratory, California 94305, USA.
Organizational Affiliation: