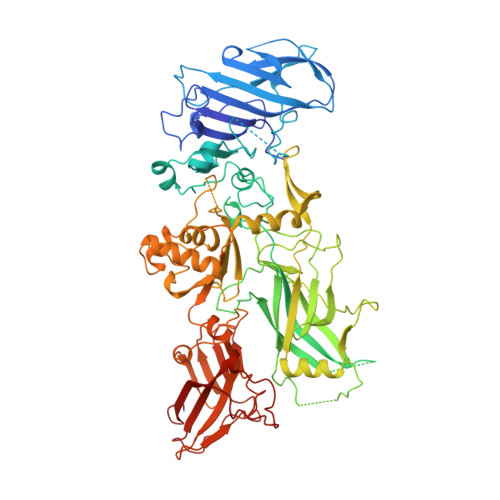
Crystal structure of the anthrax toxin protective antigen.
Petosa, C., Collier, R.J., Klimpel, K.R., Leppla, S.H., Liddington, R.C.(1997) Nature 385: 833-838
- PubMed: 9039918
- DOI: https://doi.org/10.1038/385833a0
- Primary Citation of Related Structures:
1ACC - PubMed Abstract:
Protective antigen (PA) is the central component of the three-part protein toxin secreted by Bacillus anthracis, the organism responsible for anthrax. After proteolytic activation on the host cell surface, PA forms a membrane-inserting heptamer that translocates the toxic enzymes, oedema factor and lethal factor, into the cytosol. PA, which has a relative molecular mass of 83,000 (M(r) 83K), can also translocate heterologous proteins, and is being evaluated for use as a general protein delivery system. Here we report the crystal structure of monomeric PA at 2.1 A resolution and the water-soluble heptamer at 4.5 A resolution. The monomer is organized mainly into antiparallel beta-sheets and has four domains: an amino-terminal domain (domain 1) containing two calcium ions and the cleavage site for activating proteases; a heptamerization domain (domain 2) containing a large flexible loop implicated in membrane insertion; a small domain of unknown function (domain 3); and a carboxy-terminal receptor-binding domain (domain 4). Removal of a 20K amino-terminal fragment from domain 1 allows the assembly of the heptamer, a ring-shaped structure with a negatively charged lumen, and exposes a large hydrophobic surface for binding the toxic enzymes. We propose a model of pH-dependent membrane insertion involving the formation of a porin-like, membrane-spanning beta-barrel.
- Biochemistry Department, University of Leicester, UK.
Organizational Affiliation: