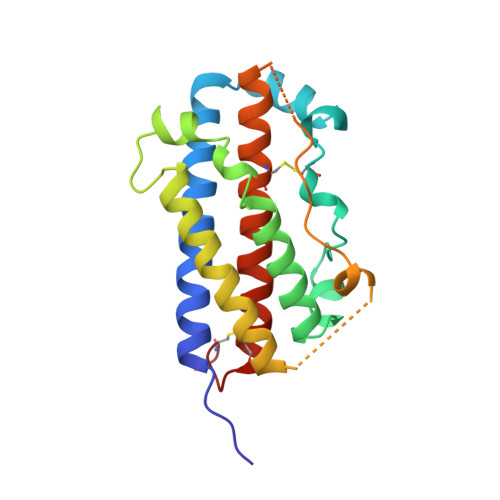
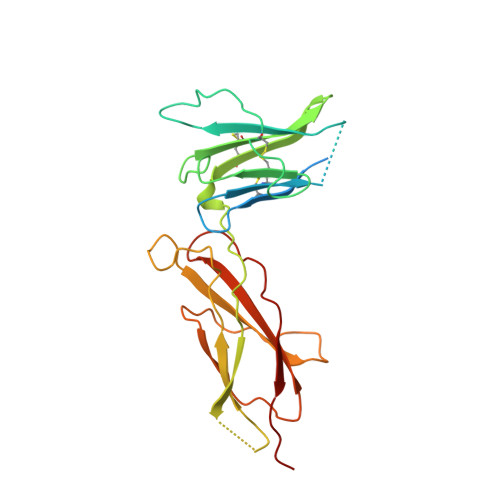
Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity.
Clackson, T., Ultsch, M.H., Wells, J.A., de Vos, A.M.(1998) J Mol Biology 277: 1111-1128
- PubMed: 9571026
- DOI: https://doi.org/10.1006/jmbi.1998.1669
- Primary Citation of Related Structures:
1A22 - PubMed Abstract:
The designed G120R mutant of human growth hormone (hGH) is an antagonist and can bind only one molecule of the growth hormone receptor. We have determined the crystal structure of the 1:1 complex between this mutant and the receptor extracellular domain (hGHbp) at 2.6 A resolution, and used it to guide a detailed survey of the structural and functional basis for hormone-receptor recognition. The overall structure of the complex is very similar to the equivalent portion of the 1:2 complex, showing that formation of the active complex does not involve major conformational changes. However, a segment involved in receptor-receptor interactions in the 1:2 complex is disordered in this structure, suggesting that its productive conformation is stabilized by receptor dimerization. The hormone binding site of the receptor comprises a central hydrophobic patch dominated by Trp104 and Trp169, surrounded by a hydrophilic periphery containing several well-ordered water molecules. Previous alanine scanning showed that the hydrophobic "hot spot" confers most of the binding energy. The new structural data, coupled with binding and kinetic analysis of further mutants, indicate that the hot spot is assembled cooperatively and that many residues contribute indirectly to binding. Several hydrophobic residues serve to orient the key tryptophan residues; kinetic analysis suggests that Pro106 locks the Trp104 main-chain into a required conformation. The electrostatic contacts of Arg43 to hGH are less important than the intramolecular packing of its alkyl chain with Trp169. The true functional epitope that directly contributes binding energy may therefore comprise as few as six side-chains, participating mostly in alkyl-aromatic stacking interactions. Outside the functional epitope, multiple mutation of residues to alanine resulted in non-additive increases in affinity: up to tenfold for a hepta-alanine mutant. Contacts in the epitope periphery can therefore attenuate the affinity of the central hot spot, perhaps reflecting a role in conferring specificity to the interaction.
- Department of Protein Engineering, Genentech, Inc., 460 Point San Bruno Blvd., South San Francisco, CA 94080, USA.
Organizational Affiliation: