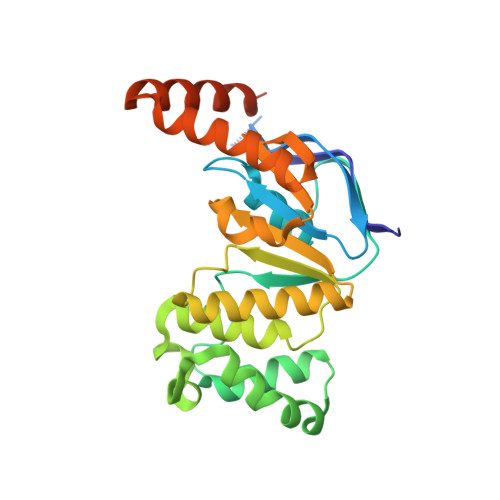
Crystal structure of the N-terminal domain of TagH reveals a potential drug targeting site.
Yang, C.S., Huang, W.C., Ko, T.P., Wang, Y.C., Wang, A.H., Chen, Y.(2020) Biochem Biophys Res Commun 536: 1-6
- PubMed: 33360015
- DOI: https://doi.org/10.1016/j.bbrc.2020.12.028
- Primary Citation of Related Structures:
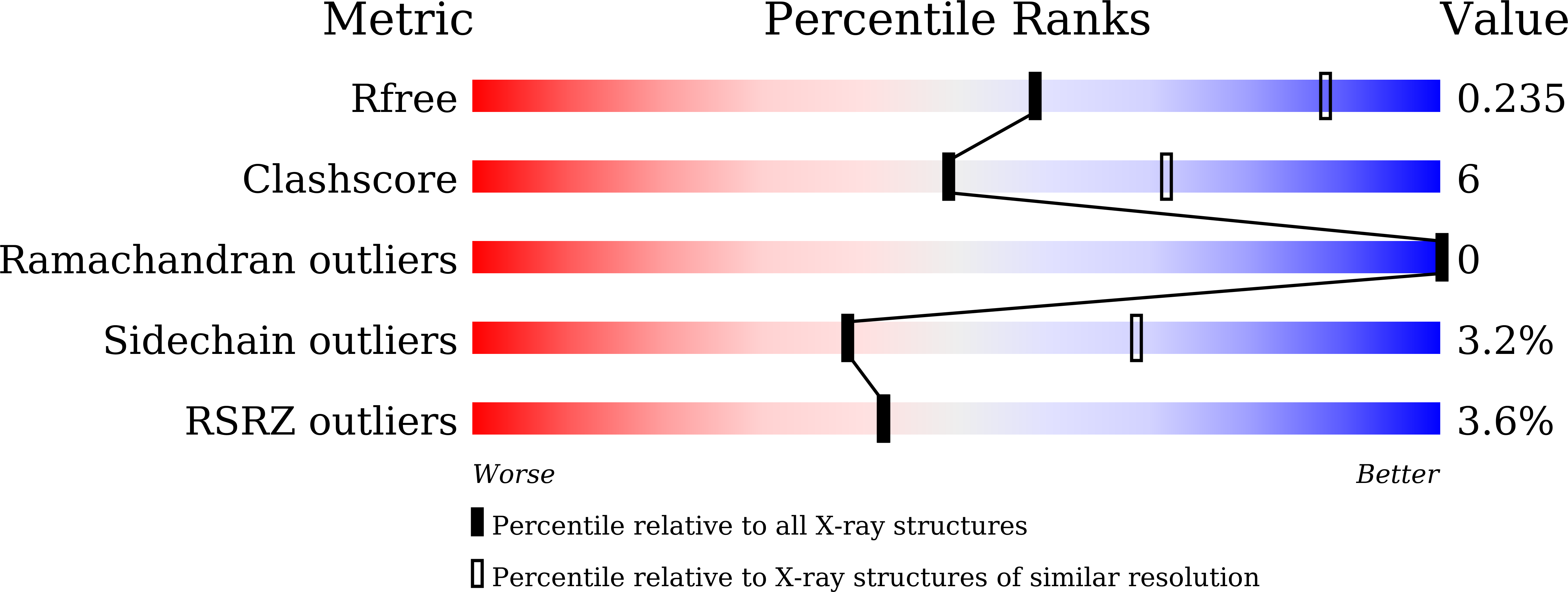
7DD0 - PubMed Abstract:
Bacterial wall teichoic acids (WTAs) are synthesized intracellularly and exported by a two-component transporter, TagGH, comprising the transmembrane and ATPase subunits TagG and TagH. Here the dimeric structure of the N-terminal domain of TagH (TagH-N) was solved by single-wavelength anomalous diffraction using a selenomethionine-containing crystal, which shows an ATP-binding cassette (ABC) architecture with RecA-like and helical subdomains. Besides significant structural differences from other ABC transporters, a prominent patch of positively charged surface is seen in the center of the TagH-N dimer, suggesting a potential binding site for the glycerol phosphate chain of WTA. The ATPase activity of TagH-N was inhibited by clodronate, a bisphosphonate, in a non-competitive manner, consistent with the proposed WTA-binding site for drug targeting.
Organizational Affiliation:
Institute of New Drug Development, China Medical University, Taichung, 406, Taiwan.