A SARS-CoV-2 antibody curbs viral nucleocapsid protein-induced complement hyperactivation.
Kang, S., Yang, M., He, S., Wang, Y., Chen, X., Chen, Y.Q., Hong, Z., Liu, J., Jiang, G., Chen, Q., Zhou, Z., Zhou, Z., Huang, Z., Huang, X., He, H., Zheng, W., Liao, H.X., Xiao, F., Shan, H., Chen, S.(2021) Nat Commun 12: 2697-2697
- PubMed: 33976229
- DOI: https://doi.org/10.1038/s41467-021-23036-9
- Primary Citation of Related Structures:
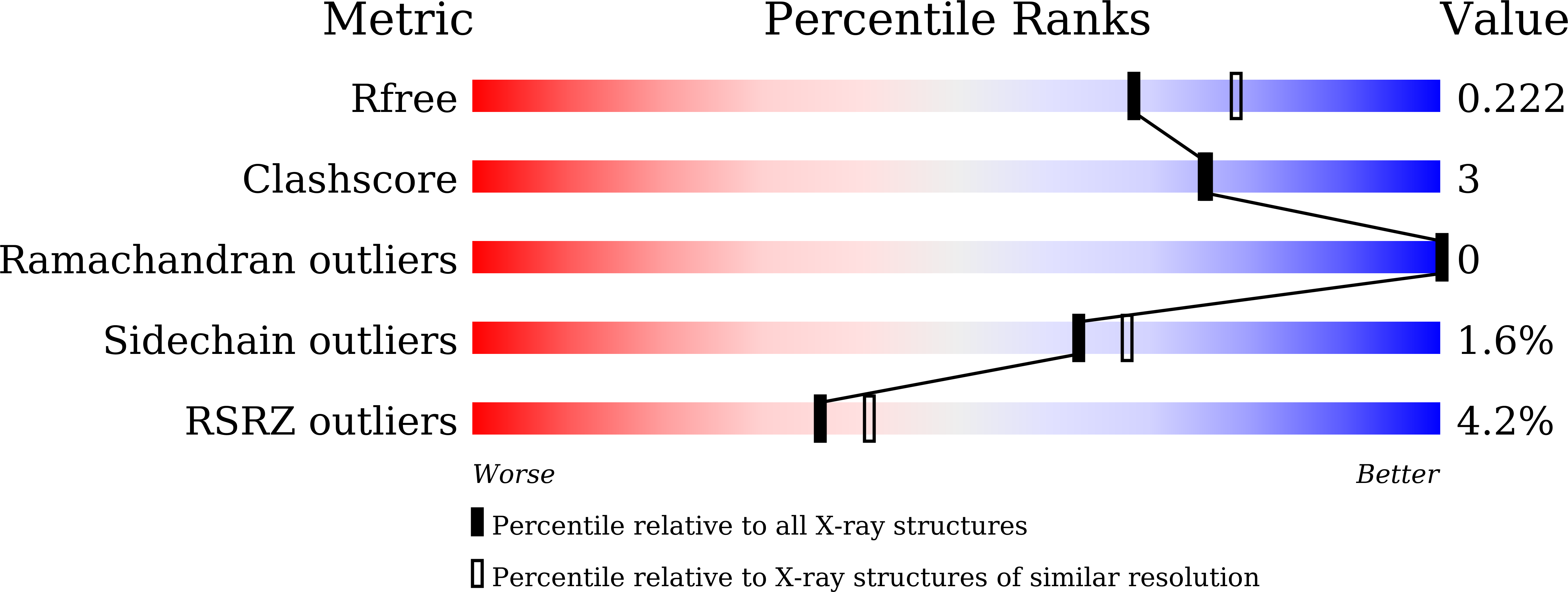
7CR5 - PubMed Abstract:
Although human antibodies elicited by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleocapsid (N) protein are profoundly boosted upon infection, little is known about the function of N-reactive antibodies. Herein, we isolate and profile a panel of 32 N protein-specific monoclonal antibodies (mAbs) from a quick recovery coronavirus disease-19 (COVID-19) convalescent patient who has dominant antibody responses to the SARS-CoV-2 N protein rather than to the SARS-CoV-2 spike (S) protein. The complex structure of the N protein RNA binding domain with the highest binding affinity mAb (nCoV396) reveals changes in the epitopes and antigen's allosteric regulation. Functionally, a virus-free complement hyperactivation analysis demonstrates that nCoV396 specifically compromises the N protein-induced complement hyperactivation, which is a risk factor for the morbidity and mortality of COVID-19 patients, thus laying the foundation for the identification of functional anti-N protein mAbs.
Organizational Affiliation:
Molecular Imaging Center, Guangdong Provincial Key Laboratory of Biomedical Imaging, The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, China.