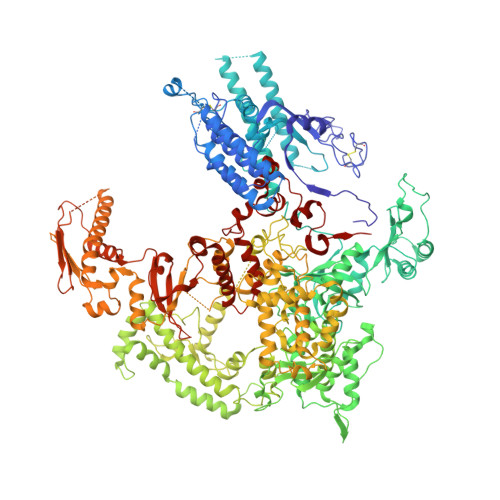
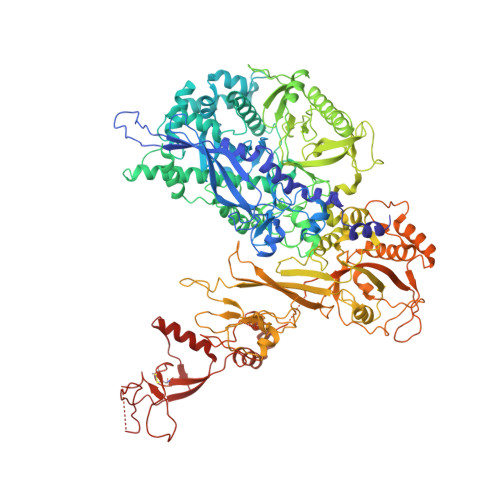
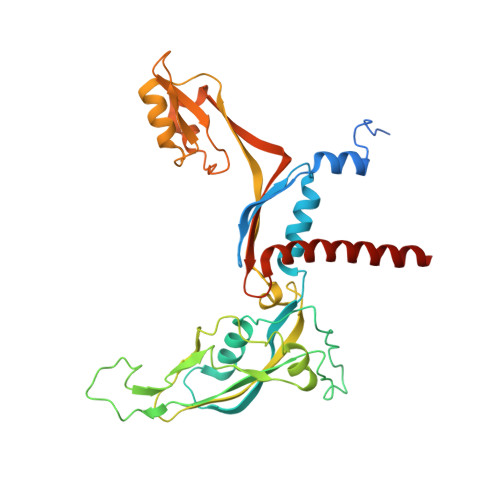
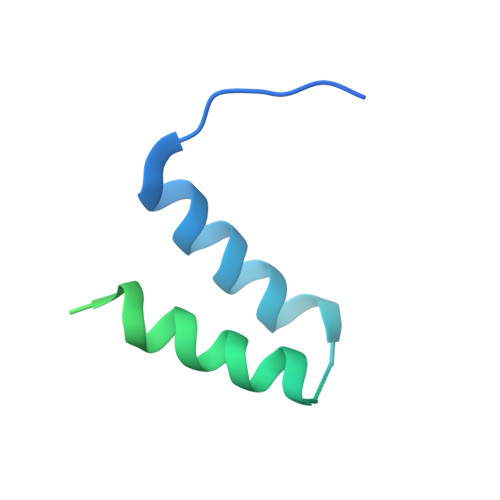
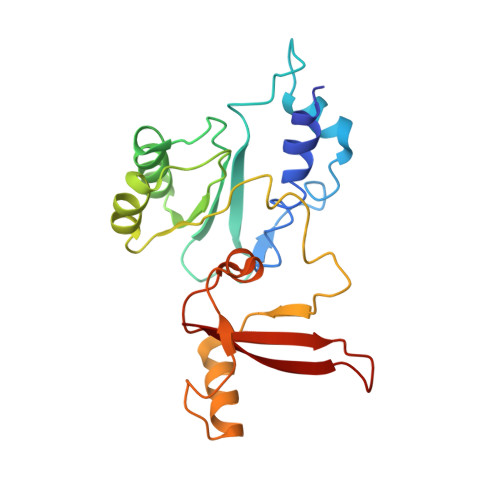
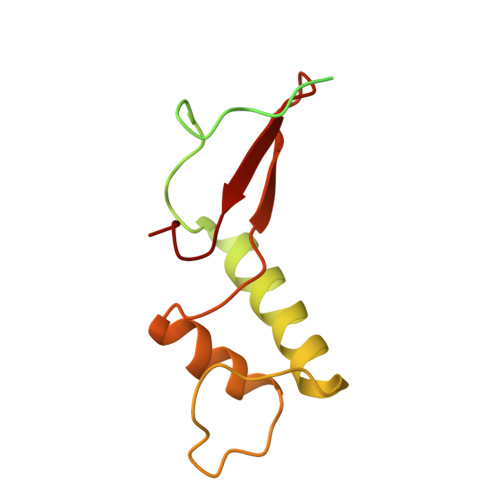
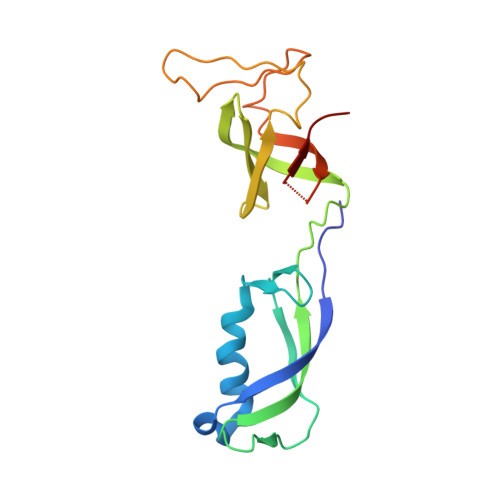
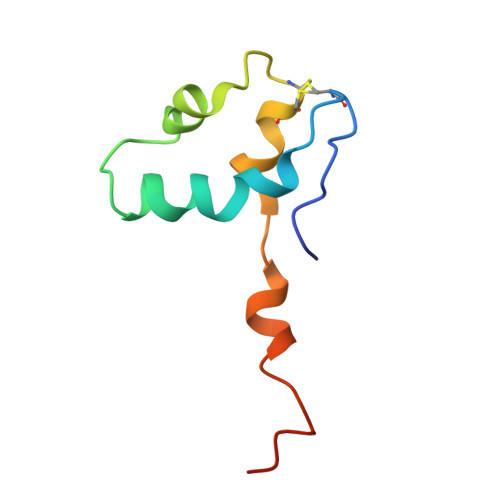
Conserved strategies of RNA polymerase I hibernation and activation.
Heiss, F.B., Daiss, J.L., Becker, P., Engel, C.(2021) Nat Commun 12: 758-758
- PubMed: 33536435
- DOI: https://doi.org/10.1038/s41467-021-21031-8
- Primary Citation of Related Structures:
7AOC, 7AOD, 7AOE - PubMed Abstract:
RNA polymerase (Pol) I transcribes the ribosomal RNA precursor in all eukaryotes. The mechanisms 'activation by cleft contraction' and 'hibernation by dimerization' are unique to the regulation of this enzyme, but structure-function analysis is limited to baker's yeast. To understand whether regulation by such strategies is specific to this model organism or conserved among species, we solve three cryo-EM structures of Pol I from Schizosaccharomyces pombe in different functional states. Comparative analysis of structural models derived from high-resolution reconstructions shows that activation is accomplished by a conserved contraction of the active center cleft. In contrast to current beliefs, we find that dimerization of the S. pombe polymerase is also possible. This dimerization is achieved independent of the 'connector' domain but relies on two previously undescribed interfaces. Our analyses highlight the divergent nature of Pol I transcription systems from their counterparts and suggest conservation of regulatory mechanisms among organisms.
- Regensburg Center for Biochemistry, University of Regensburg, Universitätsstraße 31, 93053, Regensburg, Germany.
Organizational Affiliation: