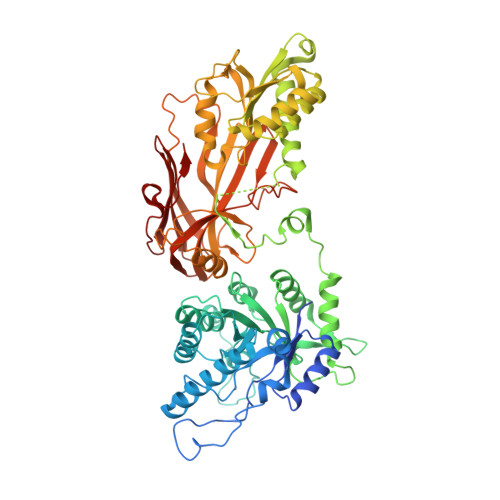
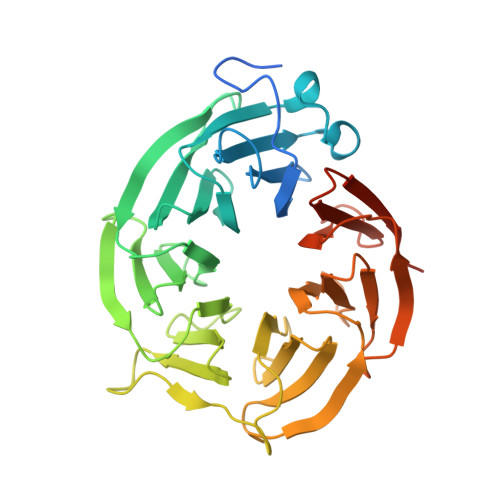
Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Palte, R.L., Schneider, S.E., Altman, M.D., Hayes, R.P., Kawamura, S., Lacey, B.M., Mansueto, M.S., Reutershan, M., Siliphaivanh, P., Sondey, C., Xu, H., Xu, Z., Ye, Y., Machacek, M.R.(2020) ACS Med Chem Lett 11: 1688-1693
- PubMed: 32944135
- DOI: https://doi.org/10.1021/acsmedchemlett.9b00525
- Primary Citation of Related Structures:
6UXX, 6UXY - PubMed Abstract:
Protein arginine methyltransferase 5 (PRMT5) belongs to a family of enzymes that regulate the posttranslational modification of histones and other proteins via methylation of arginine. Methylation of histones is linked to an increase in transcription and regulates a manifold of functions such as signal transduction and transcriptional regulation. PRMT5 has been shown to be upregulated in the tumor environment of several cancer types, and the inhibition of PRMT5 activity was identified as a potential way to reduce tumor growth. Previously, four different modes of PRMT5 inhibition were known-competing (covalently or non-covalently) with the essential cofactor S-adenosyl methionine (SAM), blocking the substrate binding pocket, or blocking both simultaneously. Herein we describe an unprecedented conformation of PRMT5 in which the formation of an allosteric binding pocket abrogates the enzyme's canonical binding site and present the discovery of potent small molecule allosteric PRMT5 inhibitors.
- Computational and Structural Chemistry, Discovery Chemistry, and Quantitative Biosciences, Merck & Co., Inc., 33 Avenue Louis Pasteur, Boston, Massachusetts 02115, United States.
Organizational Affiliation: