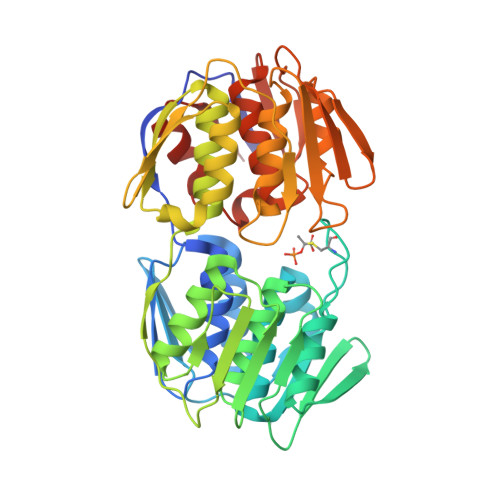
Crystal structure of MurA from Clostridium difficile in the presence of UDP-N-acetyl-alpha-D-muramic acid with modified Cys116 (S-[(1S)-1-carboxy-1-(phosphonooxy)ethyl]-L-cysteine)
Dopkins, B.J., Call, C.J., Thoden, J.B., Holden, H.M.To be published.