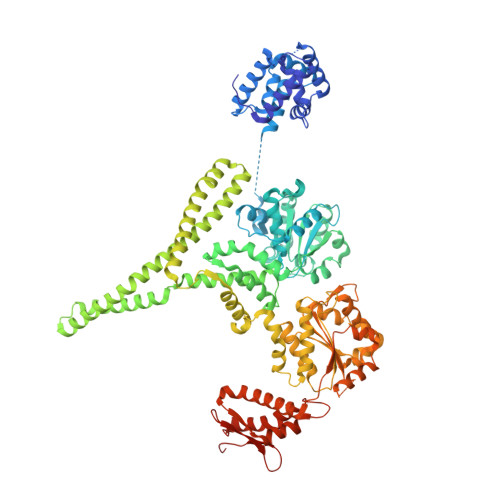
Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Michalska, K., Zhang, K., March, Z.M., Hatzos-Skintges, C., Pintilie, G., Bigelow, L., Castellano, L.M., Miles, L.J., Jackrel, M.E., Chuang, E., Jedrzejczak, R., Shorter, J., Chiu, W., Joachimiak, A.(2019) Structure 27: 449-463.e7
- PubMed: 30595457
- DOI: https://doi.org/10.1016/j.str.2018.11.001
- Primary Citation of Related Structures:
6AZY, 6D00 - PubMed Abstract:
Hsp104 is an AAA+ protein disaggregase with powerful amyloid-remodeling activity. All nonmetazoan eukaryotes express Hsp104 while eubacteria express an Hsp104 ortholog, ClpB. However, most studies have focused on Hsp104 from Saccharomyces cerevisiae and ClpB orthologs from two eubacterial species. Thus, the natural spectrum of Hsp104/ClpB molecular architectures and protein-remodeling activities remains largely unexplored. Here, we report two structures of Hsp104 from the thermophilic fungus Calcarisporiella thermophila (CtHsp104), a 2.70Å crystal structure and 4.0Å cryo-electron microscopy structure. Both structures reveal left-handed, helical assemblies with all domains clearly resolved. We thus provide the highest resolution and most complete view of Hsp104 hexamers to date. We also establish that CtHsp104 antagonizes several toxic protein-misfolding events in vivo where S. cerevisiae Hsp104 is ineffective, including rescue of TDP-43, polyglutamine, and α-synuclein toxicity. We suggest that natural Hsp104 variation is an invaluable, untapped resource for illuminating therapeutic disaggregases for fatal neurodegenerative diseases.
Organizational Affiliation:
Midwest Center for Structural Genomics, Argonne National Laboratory, Argonne, IL 60439, USA; Structural Biology Center, Biosciences Division, Argonne National Laboratory, Argonne, IL 60439, USA; Center for Structural Genomics of Infectious Diseases, Consortium for Advanced Science and Engineering, University of Chicago, Chicago, IL 60637, USA.