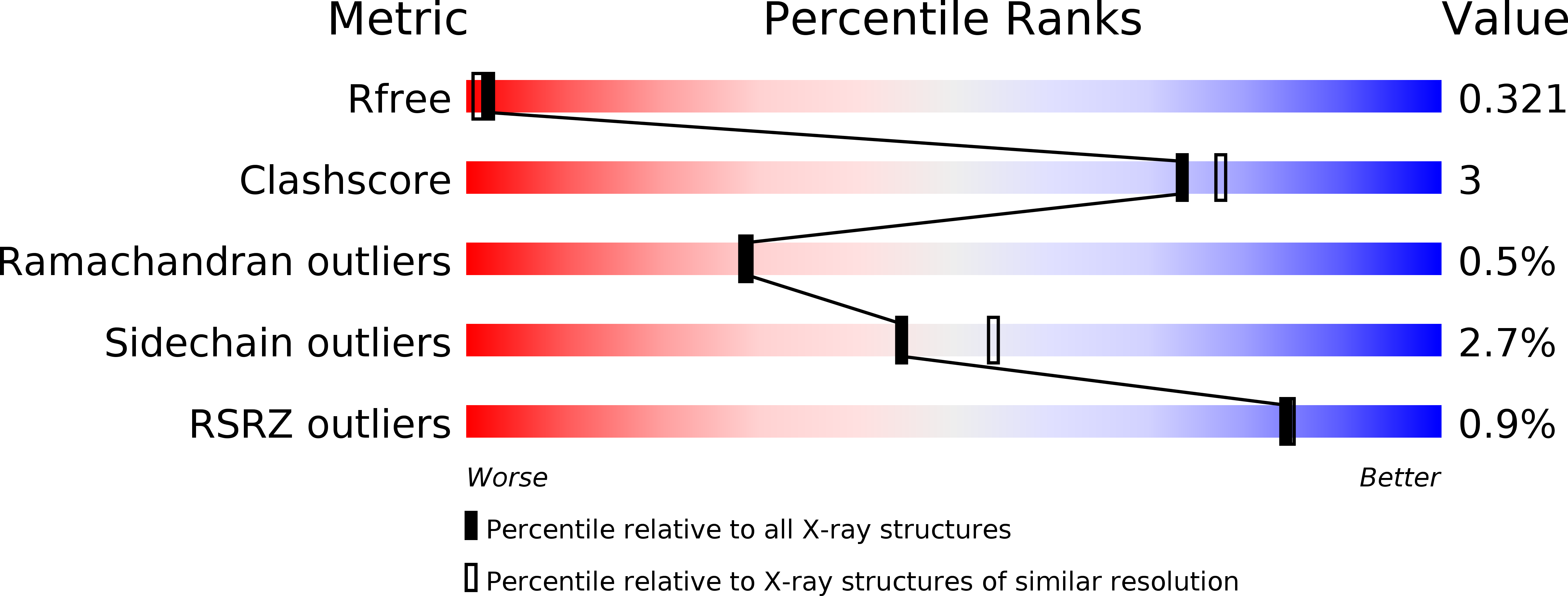
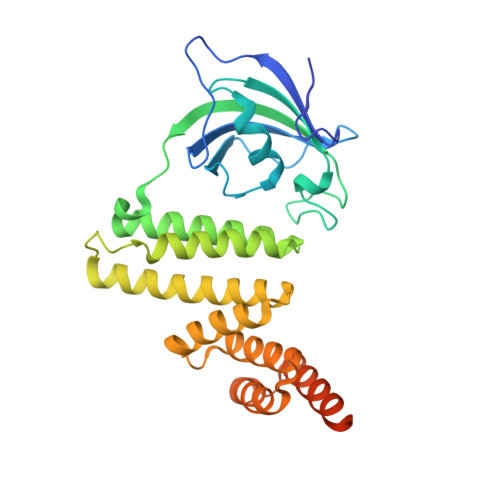
The structure of FKBP38 in complex with the MEEVD tetratricopeptide binding-motif of Hsp90.
Blundell, K.L., Pal, M., Roe, S.M., Pearl, L.H., Prodromou, C.(2017) PLoS One 12: e0173543-e0173543
- PubMed: 28278223
- DOI: https://doi.org/10.1371/journal.pone.0173543
- Primary Citation of Related Structures:
5MGX - PubMed Abstract:
Tetratricopeptide (TPR) domains are known protein interaction domains. We show that the TPR domain of FKBP8 selectively binds Hsp90, and interactions upstream of the conserved MEEVD motif are critical for tight binding. In contrast FKBP8 failed to bind intact Hsp70. The PPIase domain was not essential for the interaction with Hsp90 and binding was completely encompassed by the TPR domain alone. The conformation adopted by Hsp90 peptides, containing the conserved MEEVD motif, in the crystal structure were similar to that seen for the TPR domains of CHIP, AIP and Tah1. The carboxylate clamp interactions with bound Hsp90 peptide were a critical component of the interaction and mutation of Lys 307, involved in the carboxylate clamp, completely disrupted the interaction with Hsp90. FKBP8 binding to Hsp90 did not substantially influence its ATPase activity.
Organizational Affiliation:
Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Falmer, Brighton BN1 9RQ, England.