Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
Laponogov, I., Veselkov, D.A., Pan, X.-S., Selvarajah, J., Crevel, I.M.-T., Fisher, L.M., Sanderson, M.R.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA gyrase subunit A | A, C [auth B] | 499 | Streptococcus pneumoniae | Mutation(s): 0 Gene Names: gyrA, BM52_0349 EC: 5.99.1.3 (PDB Primary Data), 5.6.2.2 (UniProt) | 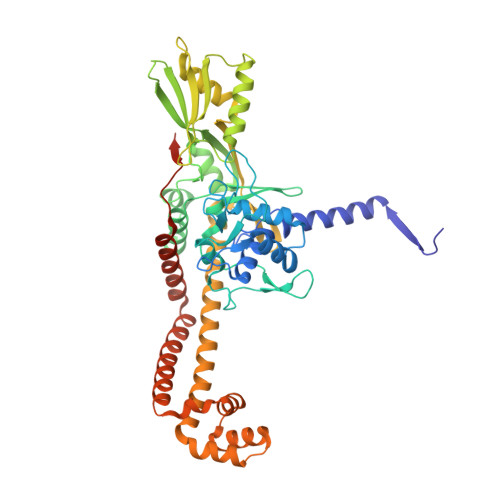 |
UniProt | |||||
Find proteins for Q8DPM2 (Streptococcus pneumoniae (strain ATCC BAA-255 / R6)) Explore Q8DPM2 Go to UniProtKB: Q8DPM2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DPM2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA gyrase subunit B | B [auth C], D | 269 | Streptococcus pneumoniae | Mutation(s): 0 Gene Names: gyrB, BM52_1967, DJ38_04430 EC: 5.99.1.3 (PDB Primary Data), 5.6.2.2 (UniProt) | 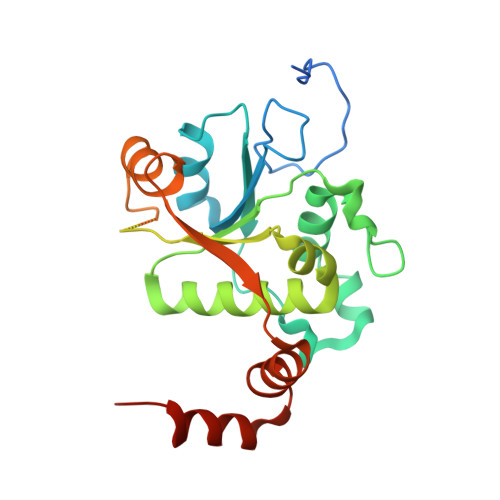 |
UniProt | |||||
Find proteins for P0A4M0 (Streptococcus pneumoniae (strain ATCC BAA-255 / R6)) Explore P0A4M0 Go to UniProtKB: P0A4M0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A4M0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| Symmetrized E-site DNA | E, H [auth G] | 15 | Escherichia coli | 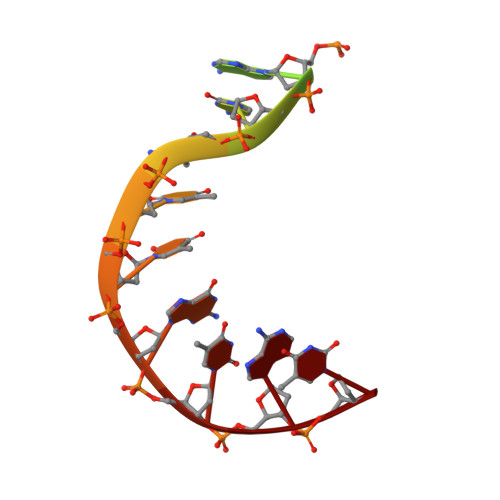 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| Symmetrized E-site DNA | F, G [auth H] | 19 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MFX Query on MFX | I [auth F], J [auth H] | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid C21 H24 F N3 O4 FABPRXSRWADJSP-MEDUHNTESA-N |  | ||
| MG Query on MG | K [auth H], L [auth G] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 92.03 | α = 90 |
| b = 94.95 | β = 90 |
| c = 274.29 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| xia2 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council | United Kingdom | BB/K010069/1 |