Structures of MauG in complex with quinol and quinone MADH.
Yukl, E.T., Jensen, L.M., Davidson, V.L., Wilmot, C.M.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 738-743
- PubMed: 23832199
- DOI: https://doi.org/10.1107/S1744309113016539
- Primary Citation of Related Structures:
4K3I - PubMed Abstract:
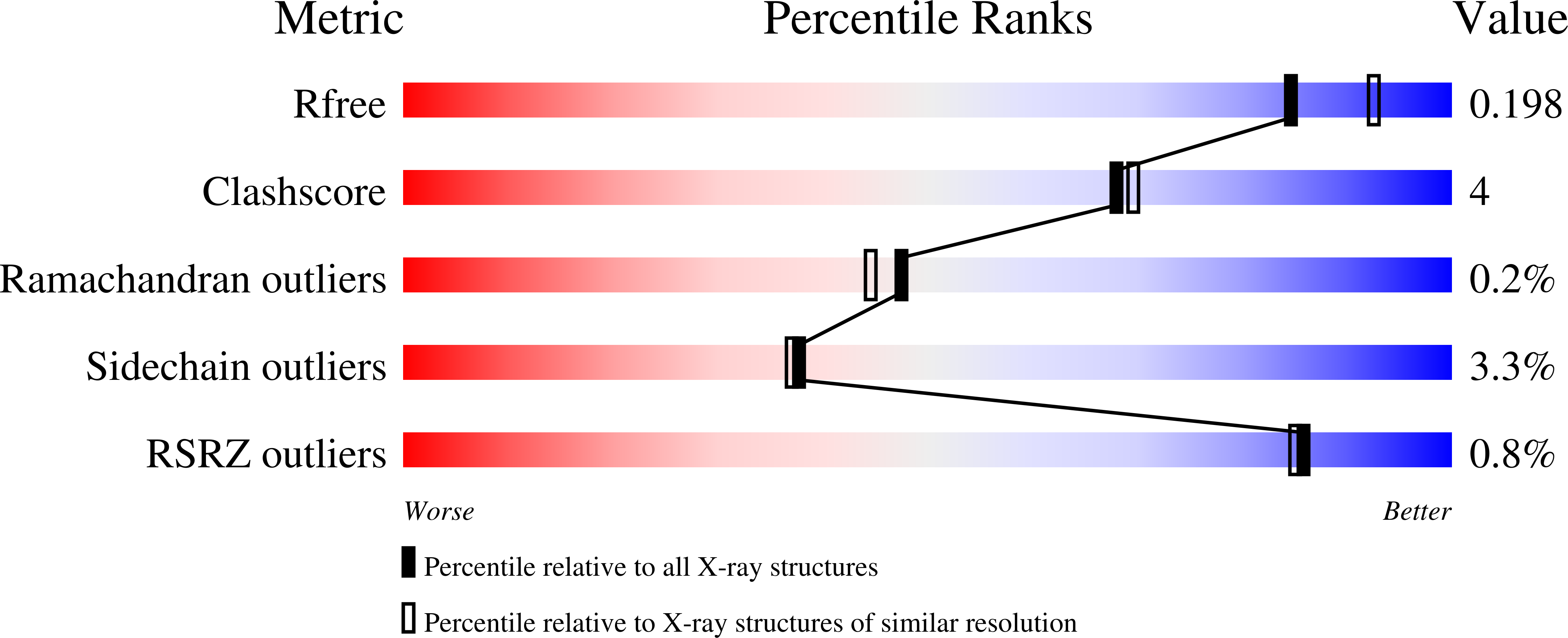
MauG has been cocrystallized with methylamine dehydrogenase (MADH) with its TTQ cofactor in the o-quinol (TTQOQ) and quinone (TTQOX) forms and the structures of the resulting complexes have been solved. The TTQOQ structure crystallized in either space group P21 or C2, while the TTQOX structure crystallized in space group P1. The previously solved structure of MauG in complex with MADH bearing an incompletely formed TTQ cofactor (preMADH) also crystallized in space group P1, although with different unit-cell parameters. Despite the changes in crystal form, the structures are virtually identical, with only very minor changes at the protein-protein interface. The relevance of these structures with respect to the measured changes in affinity between MauG and various forms of MADH is discussed.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, 321 Church Street SE, Minneapolis, MN 55455, USA.