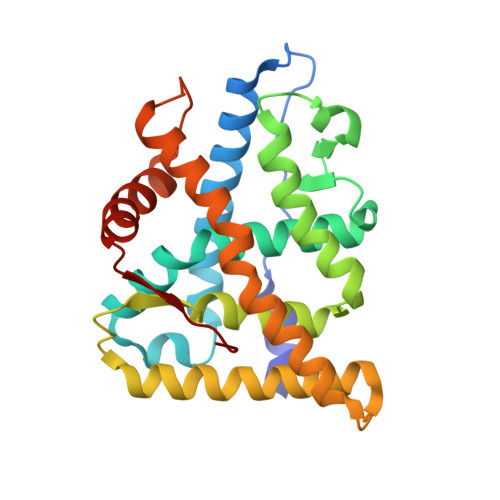
Deciphering Modern Glucocorticoid Cross-pharmacology Using Ancestral Corticosteroid Receptors.
Kohn, J.A., Deshpande, K., Ortlund, E.A.(2012) J Biol Chem 287: 16267-16275
- PubMed: 22437833
- DOI: https://doi.org/10.1074/jbc.M112.346411
- Primary Citation of Related Structures:
4E2J - PubMed Abstract:
Steroid receptors (SRs) are the largest family of metazoan transcription factors and control genes involved in development, endocrine signaling, reproduction, immunity, and cancer. The entire hormone receptor system is driven by a molecular switch triggered by the binding of small lipophilic ligands. This makes the SRs ideal pharmaceutical targets, yet even the best clinically approved synthetic steroidal agonists are prone to cross-reactivity and off-target pharmacology. The mechanism underlying this promiscuity is derived from the fact that SRs share common structural features derived from their evolutionary relationship. More often than not, rational attempts to probe SR drug selectivity via mutagenesis fail even when high quality structural and functional data are available due to the fact that important mutations often result in nonfunctional receptors. This highlights the fact that SRs suffer from instability, preventing in-depth mutational analysis and hampering crystallization of key receptor-ligand complexes. We have taken a unique approach to address this problem by using a resurrected ancestral protein to determine the structure of a previously intractable complex and identified the structural mechanisms that confer activation and selectivity for a widely used glucocorticoid, mometasone furoate. Moreover, we have identified a single residue located outside of the ligand-binding pocket that controls mometasone furoate antagonism versus agonism in the human mineralocorticoid receptor.
Organizational Affiliation:
Department of Biochemistry and the Discovery and Developmental Therapeutics Program, Winship Cancer Institute, Emory University School of Medicine, Atlanta, Georgia 30322, USA.