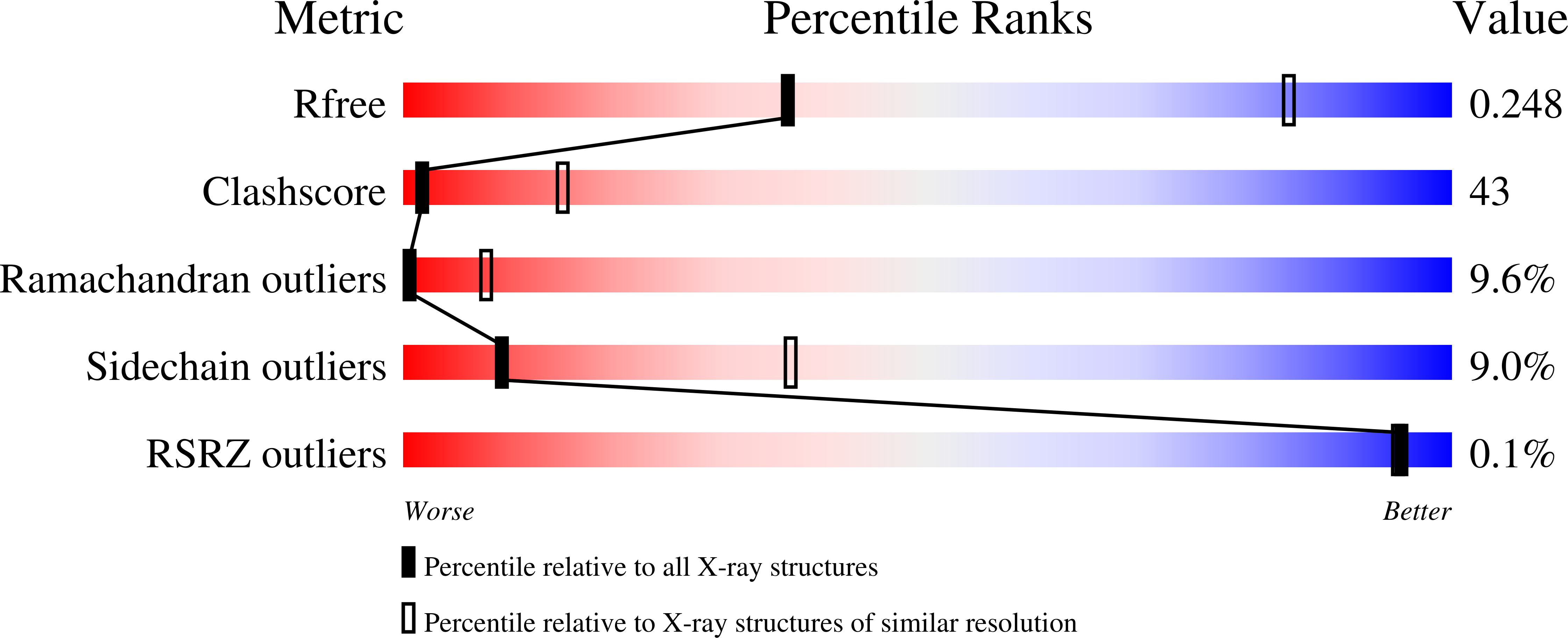
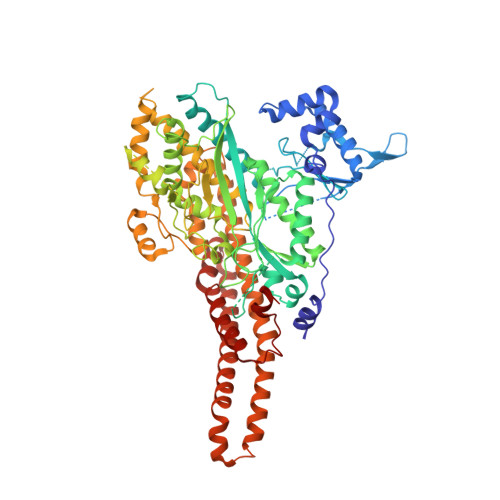
Crystal Structure of Type III Glutamine Synthetase: Surprising Reversal of the Inter-Ring Interface.
van Rooyen, J.M., Abratt, V.R., Belrhali, H., Sewell, T.(2011) Structure 19: 471-483
- PubMed: 21481771
- DOI: https://doi.org/10.1016/j.str.2011.02.001
- Primary Citation of Related Structures:
3O6X - PubMed Abstract:
Glutamine synthetases are ubiquitous, homo-oligomeric enzymes essential for nitrogen metabolism. Unlike types I and II, which are well described both structurally and functionally, the larger, type IIIs are poorly characterized despite their widespread occurrence. An understanding of the structural basis for this divergence and the implications for design of type-specific inhibitors has, therefore, been impossible. The first crystal structure of a GSIII enzyme, presented here, reveals a conservation of the GS catalytic fold but subtle differences in protein-ligand interactions suggest possible avenues for the design GSIII inhibitors. Despite these similarities, the divergence of the GSIII enzymes can be explained by differences in quaternary structure. Unexpectedly, the two hexameric rings of the GSIII dodecamer associate on the opposite surface relative to types I and II. The diversity of GS quaternary structures revealed here suggests a nonallosteric role for the evolution of the double-ringed architecture seen in all GS enzymes.
Organizational Affiliation:
Electron Microscope Unit, University of Cape Town, Rondebosch 7701, South Africa.