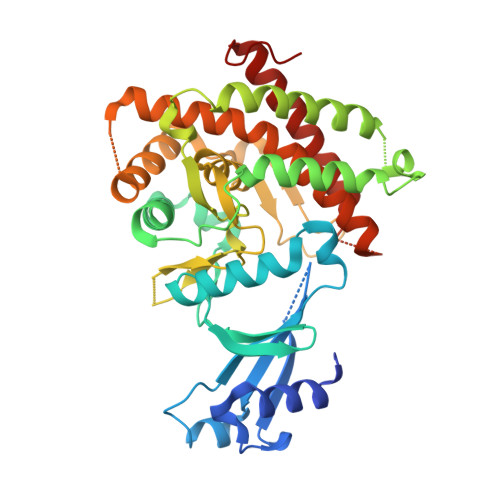
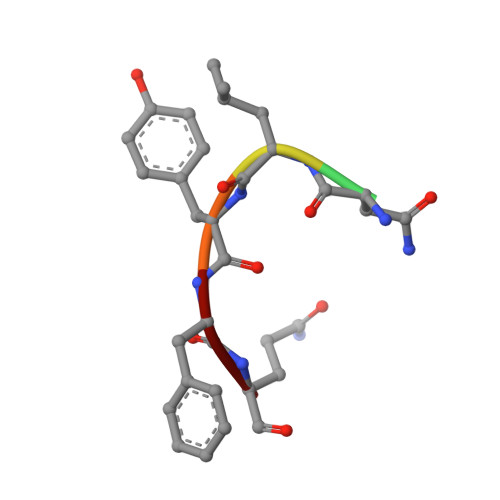
Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520.
Wernimont, A.K., Hills, T., Lew, J., Wasney, G., Senisterra, G., Kozieradzki, I., Cossar, D., Vedadi, M., Schapira, M., Bochkarev, A., Arrowsmith, C.H., Bountra, C., Weigelt, J., Edwards, A.M., Hui, R., Artz, J.D., Xiao, T.To be published.