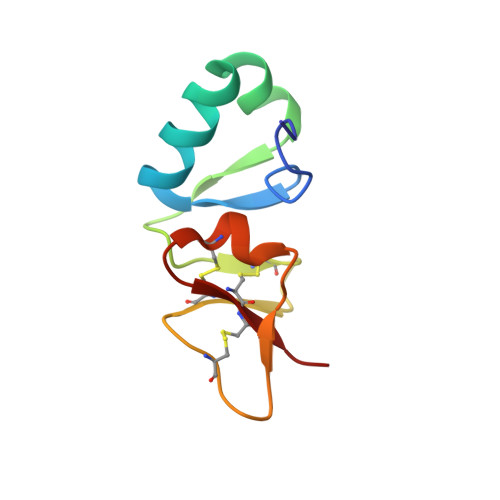A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria
Kouno, T., Fujitani, N., Mizuguchi, M., Osaki, T., Nishimura, S., Kawabata, S., Aizawa, T., Demura, M., Nitta, K., Kawano, K.(2008) Biochemistry 47: 10611-10619
- PubMed: 18785751
- DOI: https://doi.org/10.1021/bi800957n
- Primary Citation of Related Structures:
2RNG - PubMed Abstract:
Big defensin is a 79-residue peptide derived from hemocytes of the Japanese horseshoe crab. It has antimicrobial activities against Gram-positive and -negative bacteria. The amino acid sequence of big defensin can be divided into an N-terminal hydrophobic half and a C-terminal cationic half. Interestingly, the trypsin cleaves big defensin into two fragments, the N-terminal and C-terminal fragments, which are responsible for antimicrobial activity against Gram-positive and -negative bacteria, respectively. To explore the antimicrobial mechanism of big defensin, we determined the solution structure of mature big defensin and performed a titration experiment with DPC micelles. Big defensin has a novel defensin structure; the C-terminal domain adopts a beta-defensin structure, and the N-terminal domain forms a unique globular conformation. It is noteworthy that the hydrophobic N-terminal domain undergoes a conformational change in micelle solution, while the C-terminal domain remains unchanged. Here, we propose that the N-terminal domain achieves its antimicrobial activity in a novel fashion and explain that big defensin has developed a strategy different from those of other beta-defensins to suppress the growth of Gram-positive bacteria.
Organizational Affiliation:
Faculty of Pharmaceutical Sciences, University of Toyama, Toyama 930-0194, Japan.