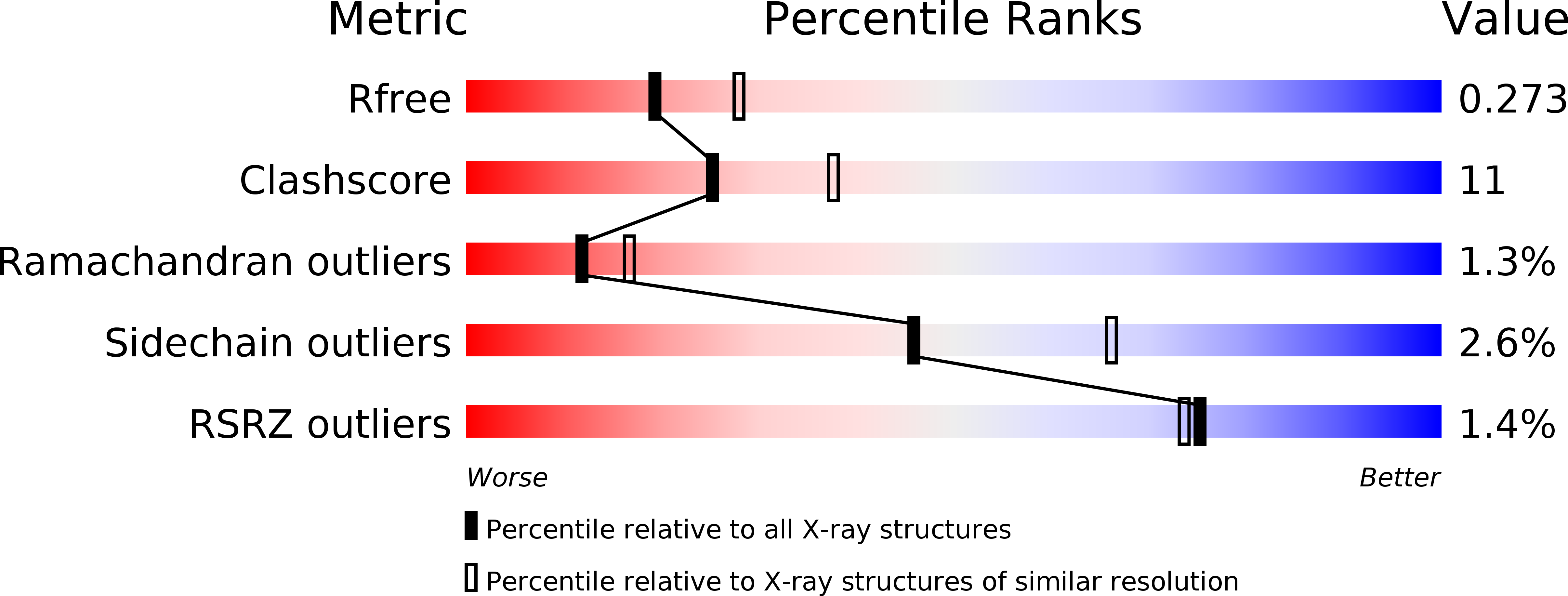
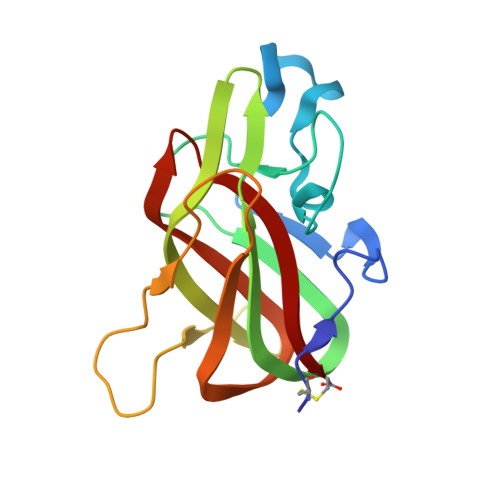
Crystal structure of the bovine lactadherin C2 domain, a membrane binding motif, shows similarity to the C2 domains of factor V and factor VIII.
Lin, L., Huai, Q., Huang, M., Furie, B., Furie, B.C.(2007) J Mol Biol 371: 717-724
- PubMed: 17583728
- DOI: https://doi.org/10.1016/j.jmb.2007.05.054
- Primary Citation of Related Structures:
2PQS - PubMed Abstract:
Lactadherin, a glycoprotein secreted by a variety of cell types, contains two EGF domains and two C domains with sequence homology to the C domains of blood coagulation proteins factor V and factor VIII. Like these proteins, lactadherin binds to phosphatidylserine (PS)-containing membranes with high affinity. We determined the crystal structure of the bovine lactadherin C2 domain (residues 1 to 158) at 2.4 A. The lactadherin C2 structure is similar to the C2 domains of factors V and VIII (rmsd of C(alpha) atoms of 0.9 A and 1.2 A, and sequence identities of 43% and 38%, respectively). The lactadherin C2 domain has a discoidin-like fold containing two beta-sheets of five and three antiparallel beta-strands packed against one another. The N and C termini are linked by a disulfide bridge between Cys1 and Cys158. One beta-turn and two loops containing solvent-exposed hydrophobic residues extend from the C2 domain beta-sandwich core. In analogy with the C2 domains of factors V and VIII, some or all of these solvent-exposed hydrophobic residues, Trp26, Leu28, Phe31, and Phe81, likely participate in membrane binding. The C2 domain of lactadherin may serve as a marker of cell surface phosphatidylserine exposure and may have potential as a unique anti-thrombotic agent.
Organizational Affiliation:
Division of Hemostasis and Thrombosis, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, MA 02215, USA.