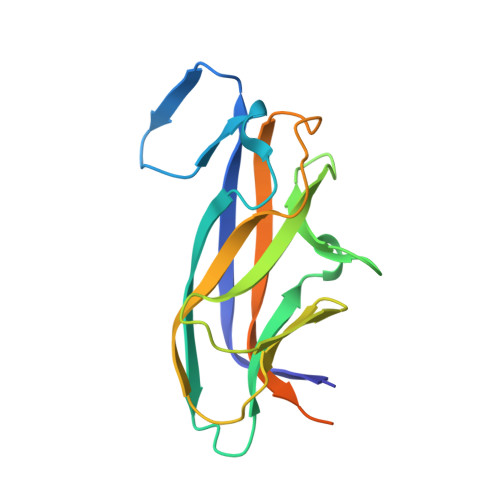
Crystal Structure and Mutational Analysis of the DaaE Adhesin of Escherichia coli.
Korotkova, N., Le Trong, I., Samudrala, R., Korotkov, K., Van Loy, C.P., Bui, A.L., Moseley, S.L., Stenkamp, R.E.(2006) J Biol Chem 281: 22367-22377
- PubMed: 16751628
- DOI: https://doi.org/10.1074/jbc.M604646200
- Primary Citation of Related Structures:
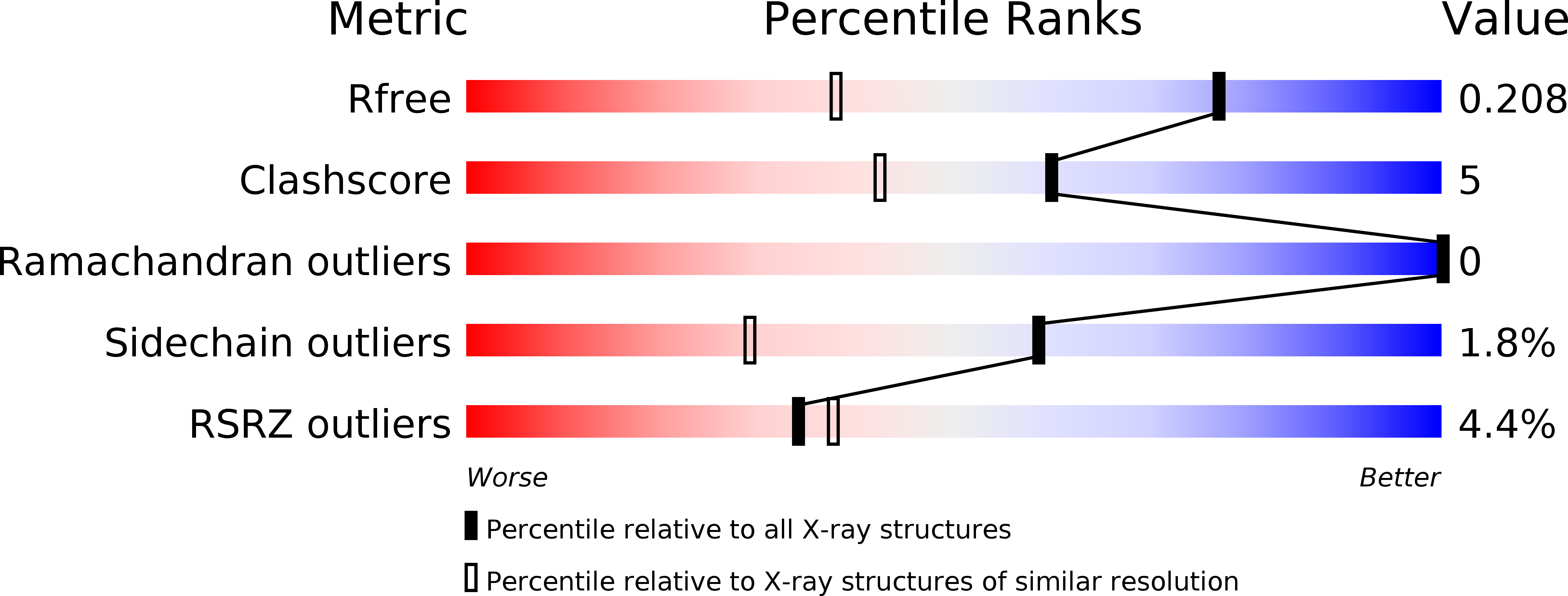
2BCM - PubMed Abstract:
DaaE is a member of the Dr adhesin family of Escherichia coli, members of which are associated with diarrhea and urinary tract infections. A receptor for Dr adhesins is the cell surface protein, decay-accelerating factor (DAF). We have carried out a functional analysis of Dr adhesins, as well as mutagenesis and crystallographic studies of DaaE, to obtain detailed molecular information about interactions of Dr adhesins with their receptors. The crystal structure of DaaE has been solved at 1.48 A resolution. Trimers of the protein are found in the crystal, as has been the case for other Dr adhesins. Naturally occurring variants and directed mutations in DaaE have been generated and analyzed for their ability to bind DAF. Mapping of the mutation sites onto the DaaE molecular structure shows that several of them contribute to a contiguous surface that is likely the primary DAF-binding site. The DAF-binding properties of purified fimbriae and adhesin proteins from mutants and variants correlated with the ability of bacteria expressing these proteins to bind to human epithelial cells in culture. DaaE, DraE, AfaE-III, and AfaE-V interact with complement control protein (CCP) domains 2-4 of DAF, and analysis of the ionic strength dependence of their binding indicates that the intermolecular interactions are highly electrostatic in nature. The adhesins AfaE-I and NfaE-2 bind to CCP-3 and CCP-4 of DAF, and electrostatic interactions contribute significantly less to these interactions. These observations are consistent with structural predictions for these Dr variants and also suggest a role for the positively charged region linking CCP-2 and CCP-3 of DAF in electrostatic Dr adhesin-DAF interactions.
Organizational Affiliation:
Department of Microbiology, University of Washington, Seattle, Washington 98195.