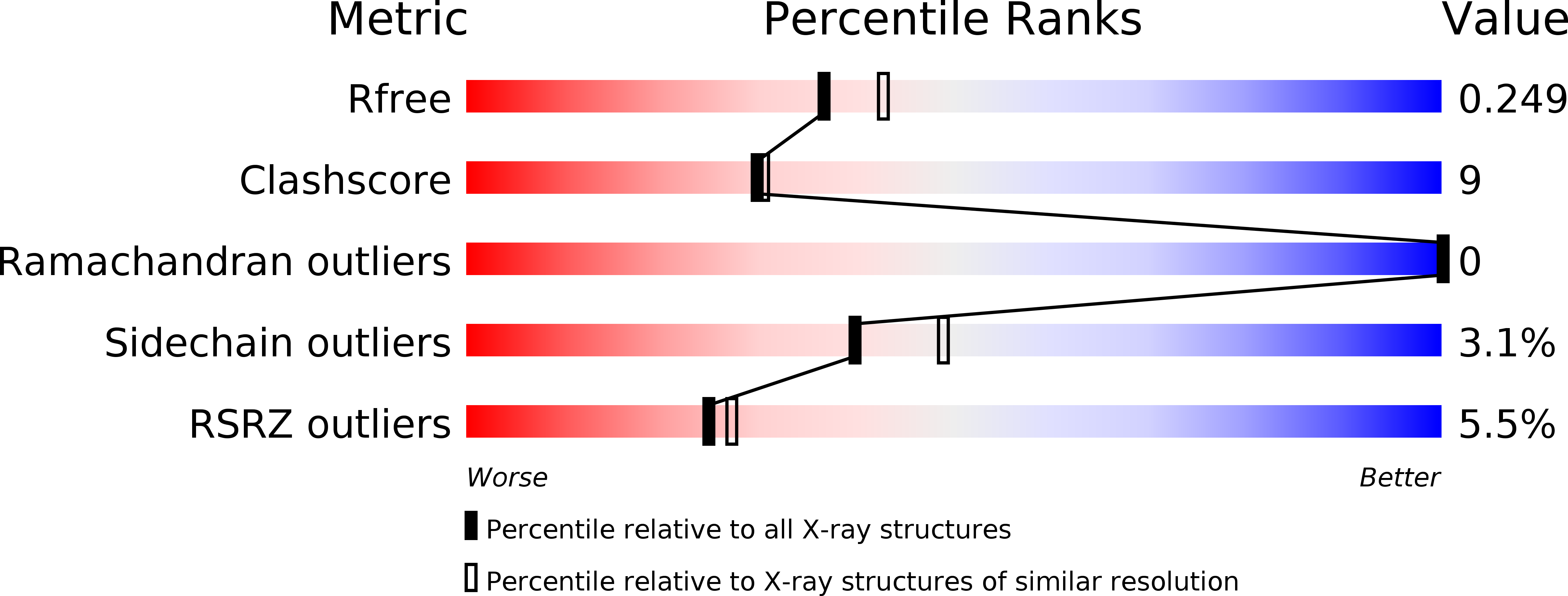
Extended Neck Regions Stabilize Tetramers of the Receptors DC-SIGN and DC-SIGNR
Feinberg, H., Guo, Y., Mitchell, D.A., Drickamer, K., Weis, W.I.(2005) J Biol Chem 280: 1327-1335
- PubMed: 15509576
- DOI: https://doi.org/10.1074/jbc.M409925200
- Primary Citation of Related Structures:
1XAR - PubMed Abstract:
The human cell surface receptors DC-SIGN (dendritic cell-specific intercellular adhesion molecule-grabbing nonintegrin) and DC-SIGNR (DC-SIGN-related) bind to oligosaccharide ligands found on human tissues as well as on pathogens including viruses, bacteria, and parasites. The extracellular portion of each receptor contains a membrane-distal carbohydrate-recognition domain (CRD) and forms tetramers stabilized by an extended neck region consisting of 23 amino acid repeats. Cross-linking analysis of full-length receptors expressed in fibroblasts confirms the tetrameric state of the intact receptors. Hydrodynamic studies on truncated receptors demonstrate that the portion of the neck of each protein adjacent to the CRD is sufficient to mediate the formation of dimers, whereas regions near the N terminus are needed to stabilize the tetramers. Some of the intervening repeats are missing from polymorphic forms of DC-SIGNR. Two different crystal forms of truncated DC-SIGNR comprising two neck repeats and the CRD reveal that the CRDs are flexibly linked to the neck, which contains alpha-helical segments interspersed with non-helical regions. Differential scanning calorimetry measurements indicate that the neck and CRDs are independently folded domains. Based on the crystal structures and hydrodynamic data, models for the full extracellular domains of the receptors have been generated. The observed flexibility of the CRDs in the tetramer, combined with previous data on the specificity of these receptors, suggests an important role for oligomerization in the recognition of endogenous glycans, in particular those present on the surfaces of enveloped viruses recognized by these proteins.
Organizational Affiliation:
Departments of Structural Biology and of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, California 94305, USA.