Crystal structure of echicetin from Echis carinatus (Indian saw-scaled viper) at 2.4A resolution.
Jasti, J., Paramasivam, M., Srinivasan, A., Singh, T.P.(2004) J Mol Biol 335: 167-176
- PubMed: 14659748
- DOI: https://doi.org/10.1016/j.jmb.2003.10.048
- Primary Citation of Related Structures:
1OZ7 - PubMed Abstract:
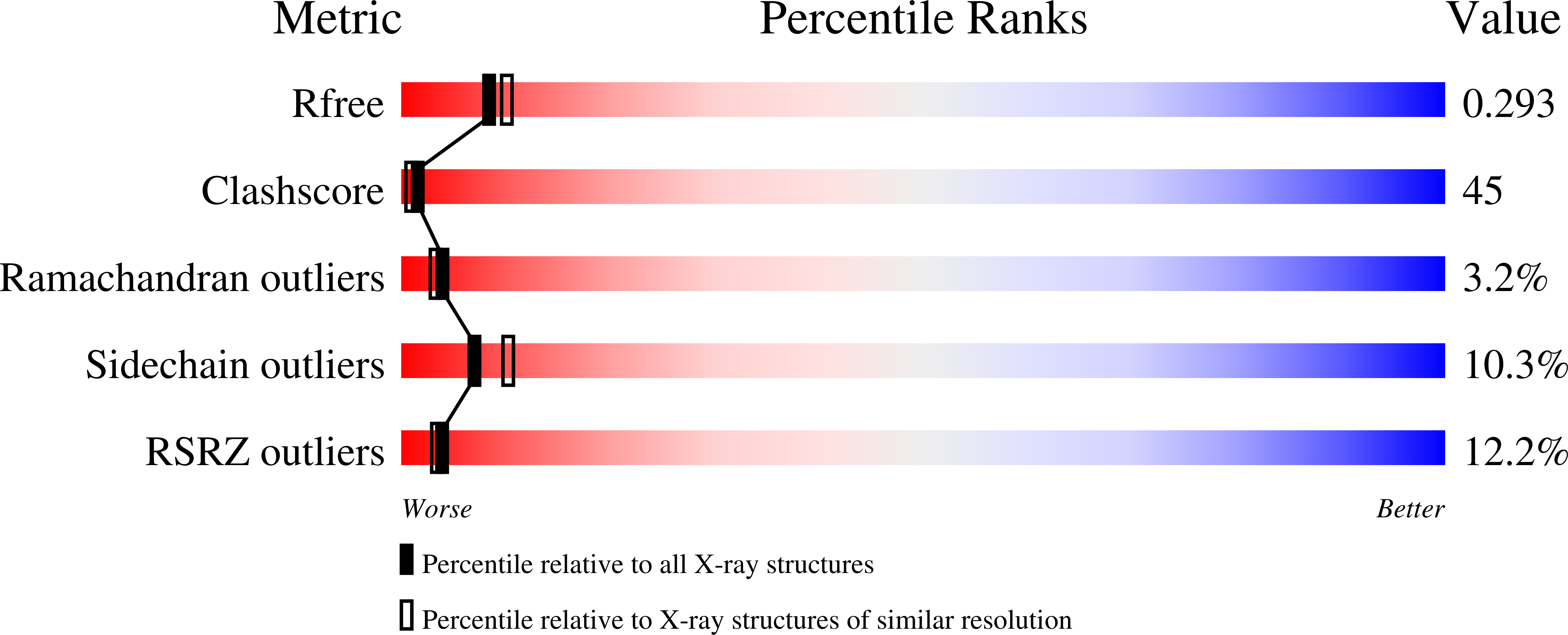
Echicetin is a heterodimeric protein from the venom of the Indian saw-scaled viper, Echis carinatus. It binds to platelet glycoprotein Ib (GPIb) and thus inhibits platelet aggregation. It has two subunits, alpha and beta, consisting of 131 and 123 amino acid residues, respectively. The two chains are linked with a disulphide bond. The level of amino acid sequence homology between two subunits is 50%. The protein was purified from the venom of E.carinatus and crystallized using ammonium sulphate as a precipitant. The crystal structure has been determined at 2.4A resolution and refined to an R-factor of 0.187. Overall dimensions of the heterodimer are approximately 80Ax35Ax35A. The backbone folds of the two subunits are similar. The central portions of the polypeptide chains of alpha and beta-subunits move into each other to form a tight dimeric association. The remaining portions of the chains of both subunits fold in a manner similar to those observed in the carbohydrate-binding domains of C-type lectins. In echicetin, the Ca(2+)-binding sites are not present, despite being topologically equivalent to other similar Ca(2+)-binding proteins of the superfamily. The residues Ser41, Glu43 and Glu47 in the calcium-binding proteins of the related family are conserved but the residues Glu126/120 are replaced by lysine at the corresponding sites in the alpha and beta-subunits.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, Ansari Nagar, 110 029, New Delhi, India.