NMR Structure of the HIV-1 Regulatory Protein Vpr
Morellet, N., Bouaziz, S., Lenoir, C., Roques, B.P.(2003) J Mol Biol 327: 215-227
- PubMed: 12614620
- DOI: https://doi.org/10.1016/s0022-2836(03)00060-3
- Primary Citation of Related Structures:
1M8L - PubMed Abstract:
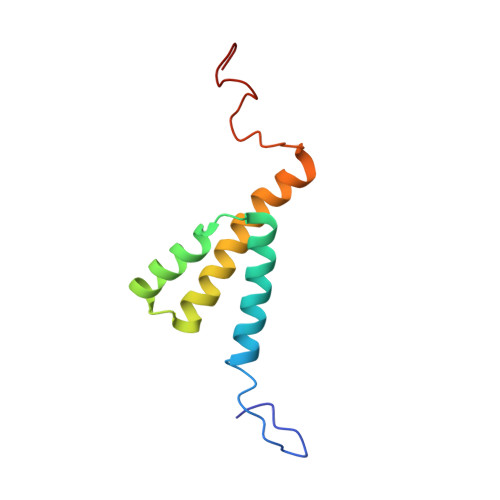
The human immunodeficiency virus type 1 (HIV-1) genome encodes a highly conserved regulatory gene product, Vpr (96 residues, 14kDa), which is incorporated into virions. In the infected cells, Vpr, expressed late in the virus cycle, is believed to function in the early phases of HIV-1 replication, such as nuclear migration of pre-integration complex, transcription of the proviral genome, viral multiplication by blocking cells in G2 phase and regulation of apoptosis phenomenon. Vpr has a critical role in long term AIDS disease by inducing infection in non-dividing cells such as monocytes and macrophages. To gain insight into the structure-function relationships of Vpr, the (1-96)Vpr protein was synthesized with 22 labeled amino acids. Its 3D structure was analyzed in the presence of CD(3)CN and in pure water at low pH and refined by restrained simulated annealing. The structure of the protein is characterized by three well-defined alpha-helices: 17-33, 38-50 and 56-77 surrounded by flexible N and C-terminal domains. In contrast to the structure obtained in the presence of TFE, the three alpha-helices are folded around a hydrophobic core constituted of Leu, Ile, Val and aromatic residues as illustrated by numerous long range NOEs. This structure accounts for the interaction of Vpr with different targets.
Organizational Affiliation:
Département de Pharmacochimie Moléculaire et Structurale, INSERM U266-CNRS FRE 2463, UFR des Sciences Pharmaceutiques et Biologiques, 4 Avenue de l'Observatoire, 75270 Paris Cedex 06, France. morellet@pharmacie.univ-paris5.fr