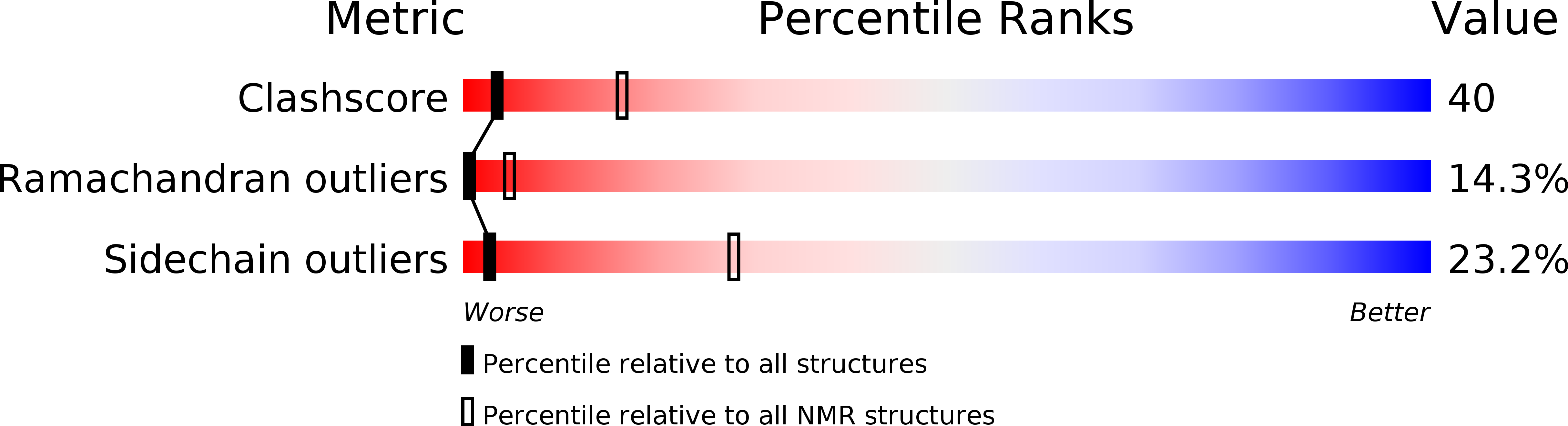Barley lipid-transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands.
Lerche, M.H., Kragelund, B.B., Bech, L.M., Poulsen, F.M.(1997) Structure 5: 291-306
- PubMed: 9032083
- DOI: https://doi.org/10.1016/s0969-2126(97)00186-x
- Primary Citation of Related Structures:
1JTB - PubMed Abstract:
. Plant nonspecific lipid-transfer proteins (nsLTPs) bind a variety of very different lipids in vitro, including phospholipids, glycolipids, fatty acids and acyl coenzyme As. In this study we have determined the structure of a nsLTP complexed with palmitoyl coenzyme A (PCoA) in order to further our understanding of the structural mechanism of the broad specificity of these proteins and its relation to the function of nsLTPs in vivo.
Organizational Affiliation:
Carlsberg Laboratorium, Kemisk Afdeling, Gamle Carlsberg Vej 10, DK-2500 Valby, Copenhagen, Denmark.