The structure of Escherichia coli heat-stable enterotoxin b by nuclear magnetic resonance and circular dichroism.
Sukumar, M., Rizo, J., Wall, M., Dreyfus, L.A., Kupersztoch, Y.M., Gierasch, L.M.(1995) Protein Sci 4: 1718-1729
- PubMed: 8528070
- DOI: https://doi.org/10.1002/pro.5560040907
- Primary Citation of Related Structures:
1EHS - PubMed Abstract:
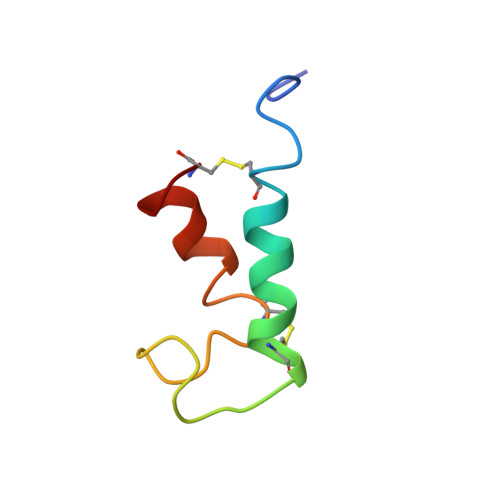
The heat-stable enterotoxin b (STb) is secreted by enterotoxigenic Escherichia coli that cause secretory diarrhea in animals and humans. It is a 48-amino acid peptide containing two disulfide bridges, between residues 10 and 48 and 21 and 36, which are crucial for its biological activity. Here, we report the solution structure of STb determined by two- and three-dimensional NMR methods. Approximate interproton distances derived from NOE data were used to construct structures of STb using distance-geometry and simulated annealing procedures. The NMR-derived structure shows that STb is helical between residues 10 and 22 and residues 38 and 44. The helical structure in the region 10-22 is amphipathic and exposes several polar residues to the solvent, some of which have been shown to be important in determining the toxicity of STb. The hydrophobic residues on the opposite face of this helix make contacts with the hydrophobic residues of the C-terminal helix. The loop region between residues 21 and 36 has another cluster of hydrophobic residues and exposes Arg 29 and Asp 30, which have been shown to be important for intestinal secretory activity. CD studies show that reduction of disulfide bridges results in a dramatic loss of structure, which correlates with loss of function. Reduced STb adopts a predominantly random-coil conformation. Chromatographic measurements of concentrations of native, fully reduced, and single-disulfide species in equilibrium mixtures of STb in redox buffers indicate that the formation of the two disulfide bonds in STb is only moderately cooperative. Similar measurements in the presence of 8 M urea suggest that the native secondary structure significantly stabilizes the disulfide bonds.
Organizational Affiliation:
Department of Chemistry, University of Massachusetts, Amherst 01003, USA.