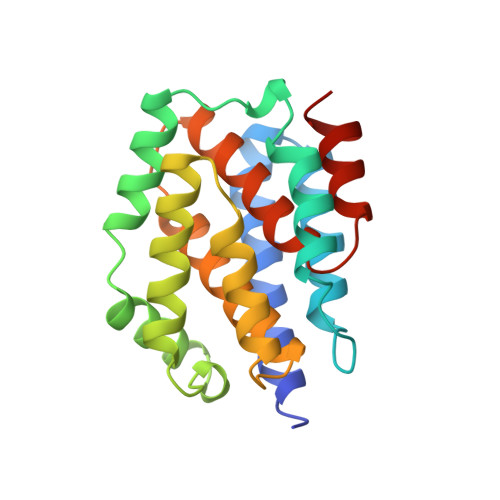
Refined structure of the pore-forming domain of colicin A at 2.4 A resolution.
Parker, M.W., Postma, J.P., Pattus, F., Tucker, A.D., Tsernoglou, D.(1992) J Mol Biol 224: 639-657
- PubMed: 1373773
- DOI: https://doi.org/10.1016/0022-2836(92)90550-4
- Primary Citation of Related Structures:
1COL - PubMed Abstract:
The E1 subgroup (E1, A, B, IA, IB, K and N) of anti-bacterial toxins called colicins is known to form voltage-dependent channels in lipid bilayers. The crystal structure of the pore-forming domain of colicin A from Escherichia coli has been refined to the diffraction limit of the crystals at 2.4 A resolution by means of molecular dynamics and restrained least-squares methods to a conventional R-factor of 0.18 for all data between 6.0 and 2.4 A resolution. The polypeptide chain of 204 amino acid residues consists of ten alpha-helices organized in a three-layer structure. The helices range in length from 9 to 23 residues with an average length of 125 residues. The packing arrangement of the helices has been analysed; the packing is different from that observed in four-helix bundle proteins. The sites of 83 water molecules have been located and refined. Analysis of the structure provides insights into the mechanism of formation of a voltage-gated channel by the protein. Although it is proposed that substantial tertiary structural changes occur during membrane insertion, the secondary structural elements remain conserved. This idea has been proposed recently for a number of other protein-membrane events and thus may have more general applicability.
Organizational Affiliation:
European Molecular Biology Laboratory, Heidelberg, Germany.