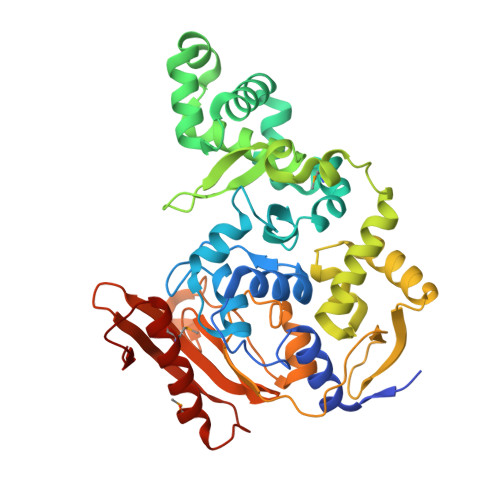
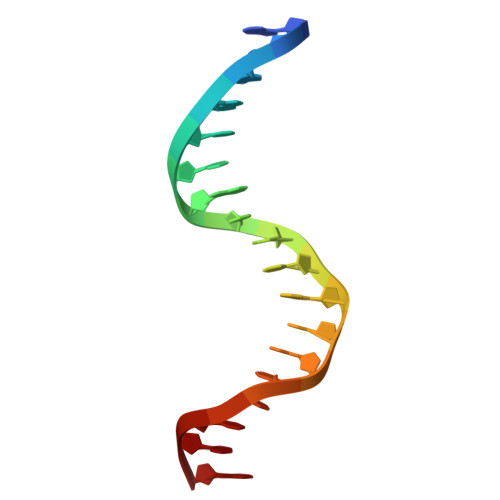
DNA binding and lesion recognition by the bacterial interstrand DNA crosslink glycosylase AlkX.
Cai 蔡毓娟, Y., Kunkle, D.E., Edinbugh, M.D., Skaar, E.P., Eichman, B.F.(2025) bioRxiv
- PubMed: 41427321
- DOI: https://doi.org/10.64898/2025.12.11.693820
- Primary Citation of Related Structures:
9ZD6 - PubMed Abstract:
Interstrand DNA crosslinks (ICLs) are a highly toxic form of DNA damage. ICL repair in both eukaryotes and bacteria involves unhooking of the two strands by specialized DNA glycosylases. We recently established that the human pathogen Acinetobacter baumannii contains an ICL glycosylase (AlkX) that facilitates pathogenesis and protects the bacteria from DNA damage and acid stress. However, the physical basis for glycosylase-catalyzed ICL unhooking is unknown. Here, we describe a crystal structure of AlkX bound to DNA representing a product of the ICL unhooking reaction. Mutational analysis of ICL unhooking in vitro and A. baumannii sensitivity to the crosslinking agent mechlorethamine enabled identification of several AlkX motifs critical for ICL repair. We also found that a genetic variant from an antibiotic-resistant strain of the human pathogen Salmonella enterica significantly reduced AlkX activity in vitro and increased A. baumannii sensitivity to DNA crosslinking. This work provides a structural basis for how bacterial ICL glycosylases recognize and repair DNA adducts and contributes additional evidence that ICL repair is important for fitness of human pathogens.
- Department of Biological Sciences, Vanderbilt University, Nashville, Tennessee 37232.
Organizational Affiliation: