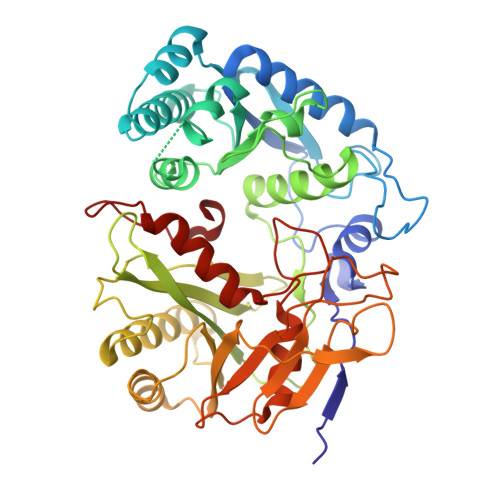
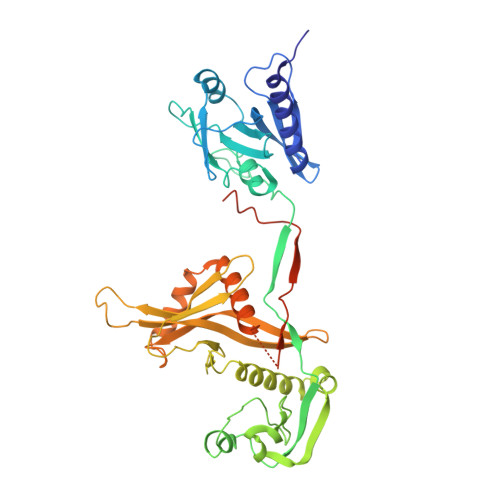
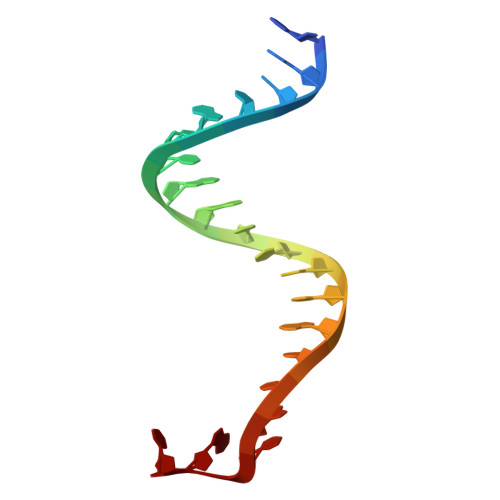
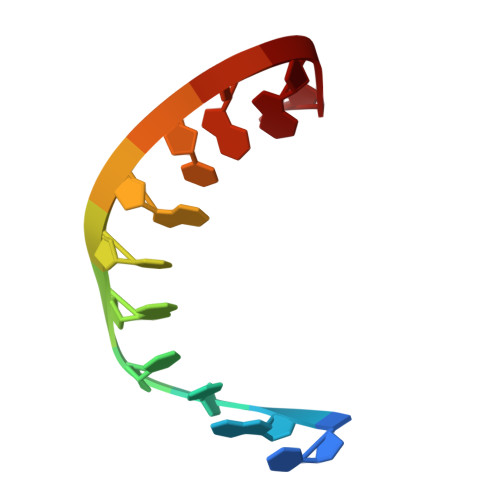
Filament assembly induced by the recognition of target DNA activates the prokaryotic Argonaute SPARDA system.
Zhang, W., Jiang, Y., Li, Y., Zhen, X., Xu, S., Xia, N.S., Li, S., Wang, X., Zheng, Q., Ouyang, S.(2026) Nat Commun 17: 115-115
- PubMed: 41495039
- DOI: https://doi.org/10.1038/s41467-025-68195-1
- Primary Citation of Related Structures:
9VX1, 9VX6 - PubMed Abstract:
The short prokaryotic Argonaute (pAgo) proteins, in conjunction with their associated effector molecules, constitute a defence mechanism in prokaryotes that protects against phage infections. The SPARDA is characterized by its collateral nuclease activity, which, upon activation through target DNA recognition, non-specifically cleaves a wide range of nucleic acid substrates. Nevertheless, the structural underpinnings of its collateral activity have remained elusive. In this study, we investigate the NbaSPARDA system from Novosphingopyxis baekryungensis and reveal that RNA-guided DNA recognition triggers the assembly of a higher-order filamentous structure. This filamentation is essential for the tetramerization of the DREN nuclease domain, which in turn facilitates the accumulation and cleavage of substrate nucleic acids. Through the determination of the gRNA-bound and RNA-DNA duplex-bound cryo-EM structures, we delineate a sequential monomer-dimer-monomer-filament transition during SPARDA activation. These insights collectively elucidate a filament-dependent activation mechanism underpinning the short pAgo-mediated immune response, which is crucial for antiviral defence.
- Key Laboratory of Microbial Pathogenesis and Interventions of Fujian Province University Provincial University, the Key Laboratory of Innate Immune Biology of Fujian Province, Biomedical Research Center of South China, College of Life Sciences, Fujian Normal University, Fuzhou, 350117, China.
Organizational Affiliation: