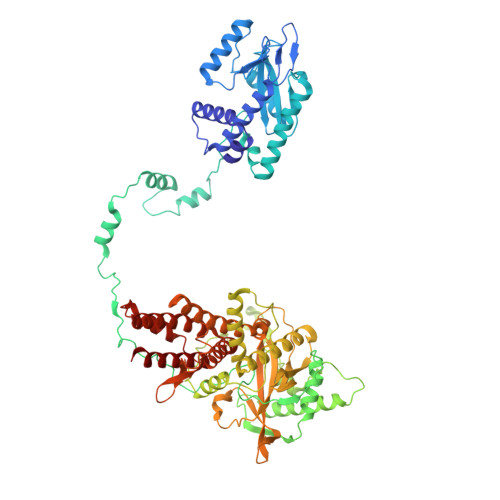
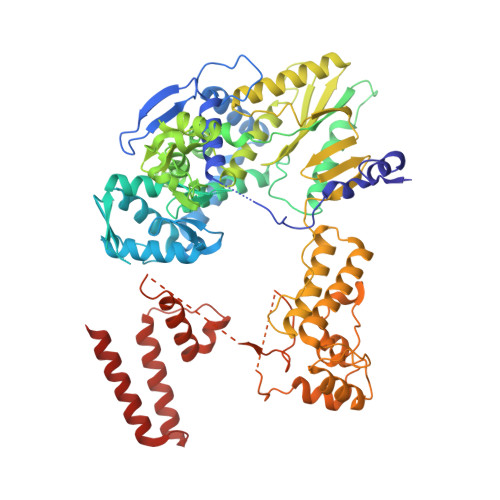
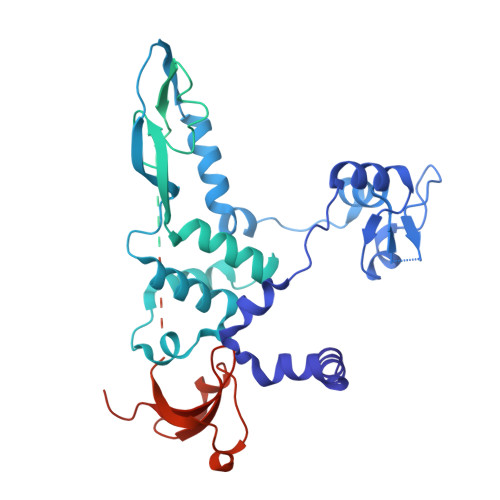
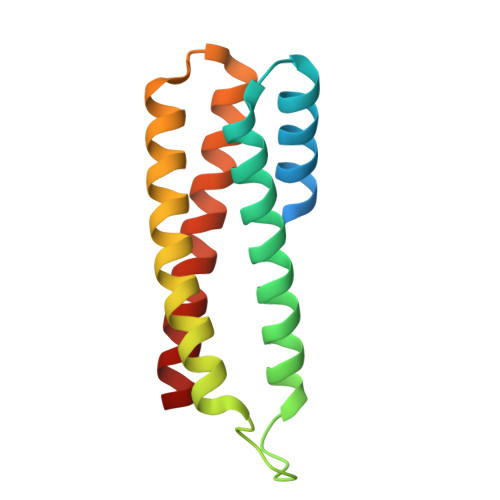
Regulatory hotspot on the influenza A virus polymerase revealed through the structure of the NEP-polymerase complex.
Rep, A., Wang, F., Chen, K.Y., Carrique, L., Sharps, J., Grimes, J.M., Fodor, E.(2026) Sci Adv 12: eaeb4073-eaeb4073
- PubMed: 41576158
- DOI: https://doi.org/10.1126/sciadv.aeb4073
- Primary Citation of Related Structures:
9RVE, 9RYC - PubMed Abstract:
Influenza A virus (IAV) transcribes and replicates its segmented RNA genome in the host nucleus within viral ribonucleoproteins (vRNPs), which are exported for virion assembly. The nuclear export protein (NEP) is essential for this process and also regulates viral RNA synthesis, implicating a direct interaction with the viral RNA polymerase. Here, we present a 2.5-Å cryogenic electron microscopy structure of NEP bound to the IAV polymerase and demonstrate that NEP alone is sufficient to promote vRNP export, with the viral matrix protein 1 enhancing export efficiency. NEP forms a four-helix bundle that binds at the interface of the PA C-terminal domain and PB1 N terminus of the polymerase. The NEP binding site at this interface overlaps with those for the host ANP32 and the C-terminal domain of RNA polymerase II, indicating that it functions as a regulatory hotspot coordinating transitions of the viral polymerase between RNA synthesis and nuclear export, revealing a critical layer of control in the IAV replication cycle.
- Sir William Dunn School of Pathology, University of Oxford, Oxford, UK.
Organizational Affiliation: