Structural insights into the antibacterial function of the Pseudomonas putida effector Tke5.
Velazquez, C., Zabala-Zearreta, M., Paredes, C., Civantos, C., Altuna-Alvarez, J., Bernal, P., Albesa-Jove, D.(2026) EMBO J
- PubMed: 41526723
- DOI: https://doi.org/10.1038/s44318-025-00689-6
- Primary Citation of Related Structures:
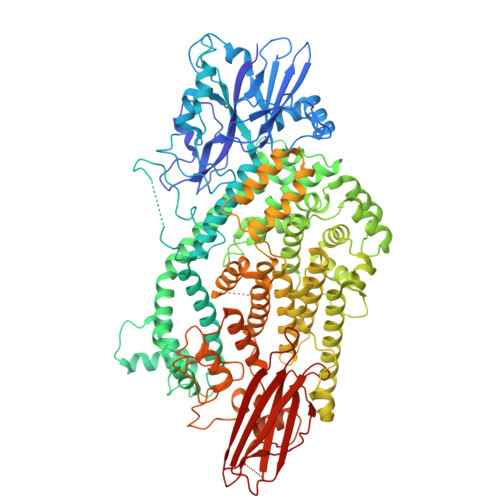
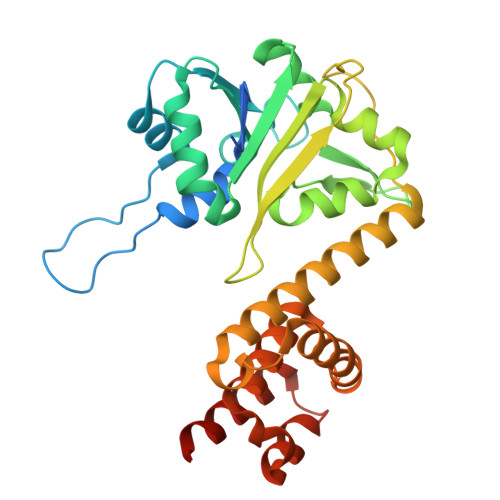
9R8G - PubMed Abstract:
Pseudomonas putida is a plant-beneficial rhizobacterium that encodes multiple type-VI secretion systems (T6SS) to outcompete phytopathogens in the rhizosphere. Among its antibacterial effectors, Tke5 (a member of the BTH_I2691 protein family) is a potent pore-forming toxin that disrupts ion homeostasis without causing considerable membrane damage. Tke5 harbours an N-terminal MIX domain, which is required for T6SS-dependent secretion in other systems. Many MIX domain-containing effectors require T6SS adaptor proteins (Tap) for secretion, but their molecular mechanisms of adaptor-effector binding remain elusive. Here, we report the 2.8 Å cryo-EM structure of the Tap3-Tke5 complex of P. putida strain KT2440, providing structural and functional insights into how effector Tke5 is recruited by its cognate adaptor protein Tap3. Functional dissection shows that the α-helical region of Tke5 is sufficient to kill intoxicated bacteria, while its β-rich region likely contributes to target membrane specificity. These findings delineate a mechanism of BTH_I2691 proteins for Tap recruitment and toxin activity, contributing to our understanding of a widespread yet understudied toxin family.
- Instituto Biofisika (CSIC, UPV/EHU), Fundación Biofísica Bizkaia/Biofisika Bizkaia Fundazioa (FBB), Leioa, 48940, Spain.
Organizational Affiliation: