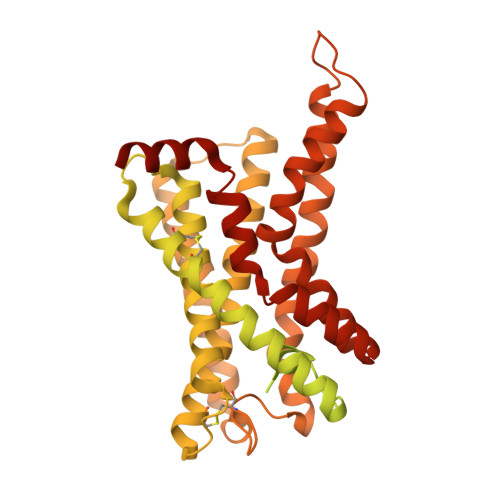
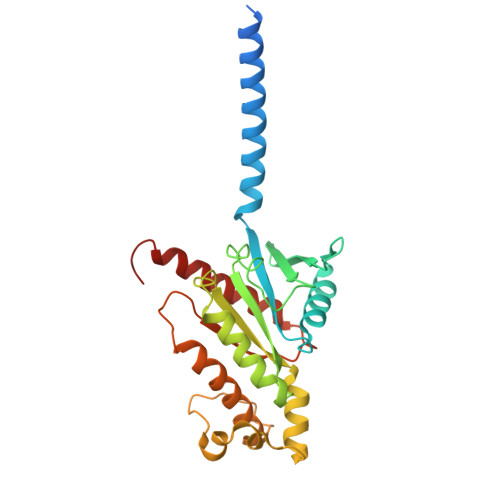
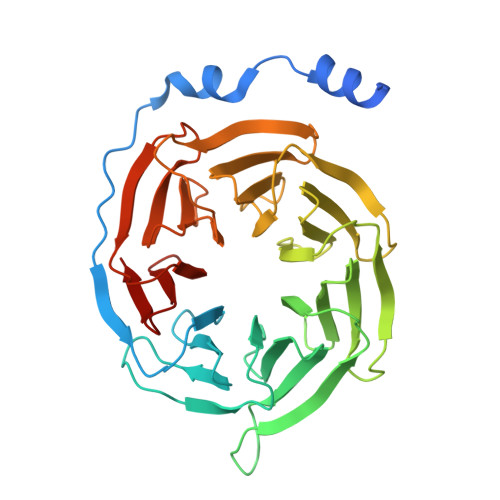
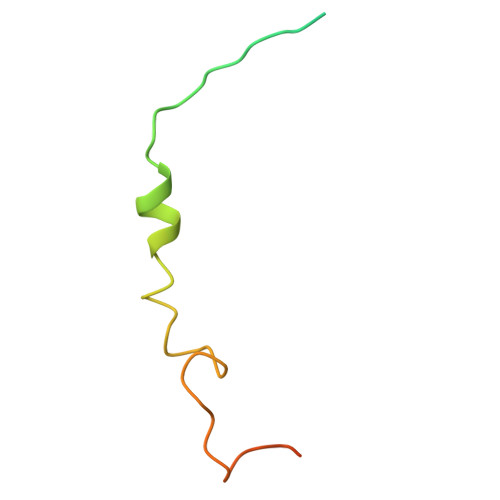
Structure of the G q -coupled adhesion receptor ADGRL4.
Chen, Q., Gusach, A., Diamante, A., Patel, J.C., Edwards, P.C., Tate, C.G., Favara, D.M.(2025) Nat Commun 17: 907-907
- PubMed: 41469374
- DOI: https://doi.org/10.1038/s41467-025-67629-0
- Primary Citation of Related Structures:
9QXL - PubMed Abstract:
Adhesion G protein-coupled receptors (aGPCRs) are a 32-member family of Class B GPCRs that have diverse cellular roles including mechanosensation, cell-fate determination, neurodevelopment, immune function and tumour biology. ADGRL4 is upregulated in the tumour microenvironment and is implicated in tumour pathogenesis across a broad range of malignancies. Inhibiting ADGRL4 is a potential therapeutic treatment for currently intractable cancers such as glioblastoma. Previous work suggested that ADGRL4 does not signal through G protein coupled pathways. However, using a sensitive bioluminescent assay, we demonstrate here that ADGRL4 couples weakly to the heterotrimeric G protein G q , whilst there is no robust coupling to other G proteins (G s , G 12 , G o ) or β-arrestin 1 or 2. We determine the cryo-EM structure of ADGRL4 coupled to a heterotrimeric G q complex to a resolution of 3.1 Å. The overall fold of ADGRL4 is similar to that of other aGPCRs, but the coupling to G q is distinct with fewer interactions between the receptor and G protein. The structure is consistent with ADGRL4 being activated by its tethered agonist and represents an important step towards the development of potential inhibitors for the treatment of multiple tumour types.
- MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge, UK.
Organizational Affiliation: