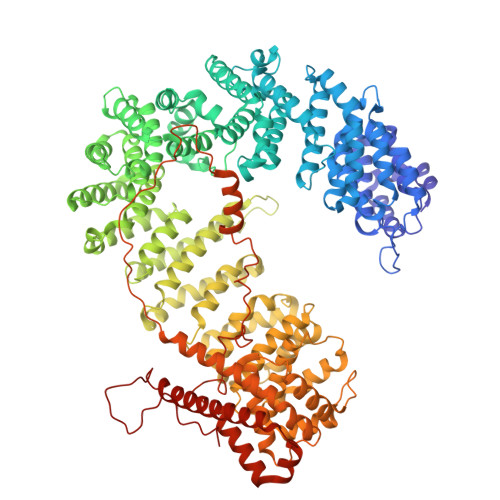
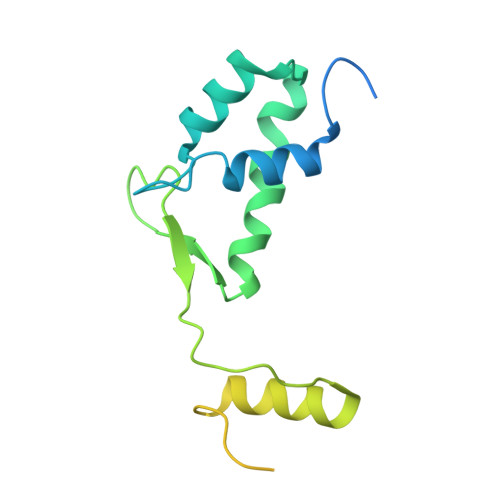
Alphafold 3-guided insights into the Importin beta : Importin7 heterodimer interaction and its binding to histone H1.
Neumann, P., Dybkov, O., Urlaub, H., Ficner, R., Dickmanns, A.(2026) Structure
- PubMed: 41529687
- DOI: https://doi.org/10.1016/j.str.2025.12.011
- Primary Citation of Related Structures:
9QEJ, 9QF0 - PubMed Abstract:
The nuclear import of H1 linker histones is facilitated by a heterodimer of the transport receptors Importinβ (Impβ) and Importin7 (Imp7). The interaction between them is mediated by a stretch of C-terminal residues of Imp7 essential also for Imp7 activation by Impβ. An Impβ:Imp7:H1 complex model was predicted by Alphafold3 and validated using cross-linking data, isothermal titration calorimetry, and pull-down experiments, providing robust support for its accuracy. This model positions the H1 globular domain within the central cavity of Imp7. Refinement of this atomic model against a published cryo-electron microscopy (cryo-EM) map demonstrated significantly improved correspondence compared to the earlier interpretation, which placed the H1 globular domain within Impβ. This enhanced structural consistency further substantiates the accuracy of the AI-driven prediction. Moreover, a detailed analysis confirmed the extended C-terminal stretch of Imp7 harboring a nucleoporin-like binding (NlB) region with two FXFG-like nucleoporin motifs interacting with the outer surface of Impβ.
- Department of Molecular Structural Biology, Institute for Microbiology and Genetics & GZMB, Georg-August-University Göttingen, 37077 Göttingen, Germany. Electronic address: pneuman2@uni-goettingen.de.
Organizational Affiliation: