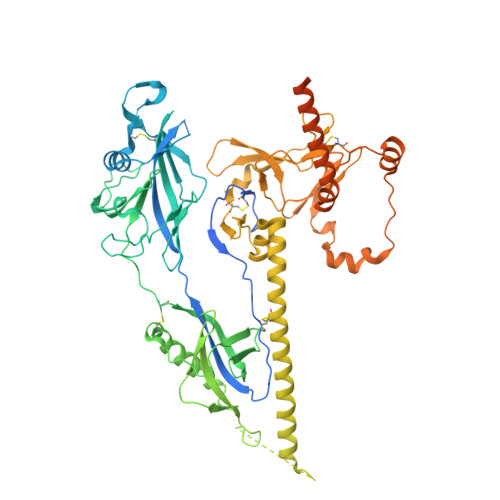
Structure and immunogenicity of an engineered soluble prefusion-stabilized EBV gB antigen.
McCool, R.S., Acreman, C.M., Powell, A.E., Picucci, S.I., Stieh, D.J., Chou, C.W., Huynh, J., Caruso, H., Park, S., O'Rear, J., Chen, J.L., Palanski, B.A., Byrne, P.O., Sponholtz, M.R., Kim, J., Ledgerwood, J.E., Weidenbacher, P.A., McLellan, J.S.(2026) Nat Commun 17: 1197-1197
- PubMed: 41484092
- DOI: https://doi.org/10.1038/s41467-025-67969-x
- Primary Citation of Related Structures:
9OAL - PubMed Abstract:
Epstein-Barr virus (EBV), the causative agent of mononucleosis, is linked to over 140,000 annual cancer-related deaths globally and increases the risk of multiple sclerosis by up to 32-fold. As a herpesvirus, EBV establishes lifelong infection, and over 90% of U.S. adults are EBV-seropositive. Despite its significant disease burden, no approved EBV vaccines or therapeutics exist. Among EBV envelope glycoproteins, the fusion protein (gB) is strictly required for epithelial and B cell infection. Here, using a combination of AlphaFold-guided modeling, rational design, and ThermoMPNN-informed optimization, we engineer a stabilized prefusion gB variant, C3-GT. This construct incorporates two inter-protomeric disulfide bonds and three cavity-filling substitutions, resulting in a melting temperature of 54 °C. Cryo-EM analysis of this construct reveals the prefusion structure of EBV gB, providing insights into the structural transitions required to adopt the postfusion conformation. Murine immunizations and depletion studies with human sera suggest a trend toward improved functional immunogenicity of C3-GT compared to postfusion gB. Collectively, these studies define engineering principles to stabilize class III fusion proteins, provide reagents to interrogate the human antibody response to EBV gB, and lay a foundation for further studies to develop EBV gB-based vaccine candidates.
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX, USA.
Organizational Affiliation: