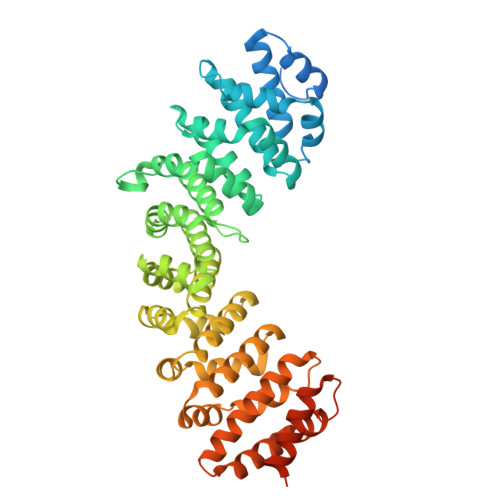
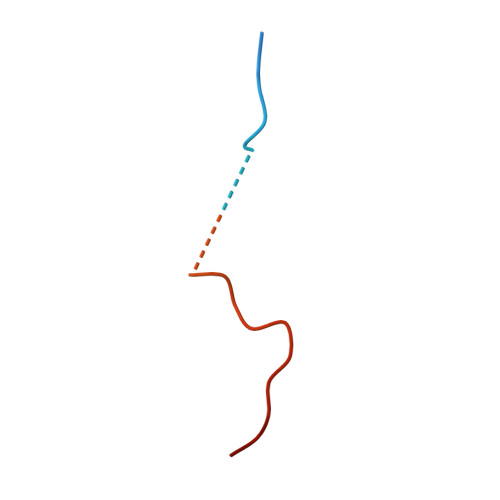
Elucidation of the binding interaction interface between AAV serotype 11 capsid protein and host nuclear import proteins.
Hoad, M., Nematollahzadeh, S., Roby, J.A., Alvisi, G., Forwood, J.K.(2025) Mol Ther Methods Clin Dev 33: 101630-101630
- PubMed: 41376776
- DOI: https://doi.org/10.1016/j.omtm.2025.101630
- Primary Citation of Related Structures:
9NZ1 - PubMed Abstract:
Adeno-associated viruses (AAVs) are widely acknowledged as versatile vectors for gene therapy due to their non-pathogenic nature, inherent capacity for tissue-specific targeting, and their potential for customizable engineering. The N terminus of the AAV capsid protein VP1 plays a pivotal role in guiding AAV capsids into the cell nucleus. However, the precise dynamics of the interaction between the VP1 protein and host nuclear transport proteins, especially across diverse AAV serotypes, remain incompletely understood. AAV11 has emerged as a promising alternative for individuals with elevated antibody titers against AAV2, and in the field of neuroscience, it has demonstrated a strong capability for mapping and manipulating neural circuits, offering the potential for treating neurological and neurodegenerative disorders. In this study, we characterize the molecular interface between AAV11 VP1 and host importin-α (IMPα). Structural and biochemical analyses reveal that the basic regions BR1 and BR3 of the VP1 N-terminal domain engage IMPα in a bipartite nuclear localization signal (NLS)-like manner. These findings provide mechanistic insight into VP1-IMPα recognition and suggest a role for these interactions in AAV11 nuclear import. While direct functional evidence is pending, this work establishes the molecular basis for VP1-host protein binding and informs future capsid engineering.
- School of Dentistry and Medical Sciences, Charles Sturt University, Wagga Wagga, NSW 2678, Australia.
Organizational Affiliation: